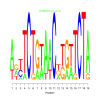
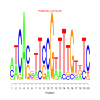
Module 199 Residual: 0.54
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0199 | v02 | 0.54 | -20.05 |
| Rv0272c | Rv0273c Transcriptional regulator, TetR family |
| Rv0274 Glyoxalase family protein | Rv0476 |
| Rv0477 POSSIBLE CONSERVED SECRETED PROTEIN | Rv1635c Possible membrane protein |
| Rv3201c ATP-dependent DNA helicase SCO5184 | Rv3526 Terminal oxygenase KshA |
| Rv3534c 4-hydroxy-2-oxovalerate aldolase (EC 4.1.3.-) | Rv3535c Acetaldehyde dehydrogenase, acetylating, (EC 1.2.1.10) in gene cluster for degradation of phenols, cresols, catechol |
| Rv3536c 2-keto-4-pentenoate hydratase (EC 4.2.1.-) | Rv3537 3-oxosteroid 1-dehydrogenase (EC 1.3.99.4) |
| Rv3538 Enoyl-CoA hydratase | Rv3568c 2,3-dihydroxybiphenyl 1,2-dioxygenase |
| Rv3569c 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase (EC 3.7.1.-) | Rv3574 Transcriptional regulator kstR (Rv3574), TetR family |
| Rv3572 |
|
GO:0005576 | extracellular region |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0070723 | response to cholesterol |
|
GO:0003824 | catalytic activity |
|
GO:0008152 | metabolic process |
|
GO:0016829 | lyase activity |
|
GO:0006725 | cellular aromatic compound metabolic process |
|
GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen |
|
GO:0005618 | cell wall |
|
GO:0034820 | 4,9-DSHA hydrolase activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0044117 | growth of symbiont in host |
|
GO:0003677 | DNA binding |



















Discussion