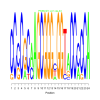
Module 242 Residual: 0.54
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0242 | v02 | 0.54 | -31.61 |
| Rv0228 Lysophospholipid acyltransferase | Rv0552 Amidohydrolase family protein |
| Rv0553 O-succinylbenzoate synthase (EC 4.2.1.113) | Rv0554 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (EC 4.2.99.20) |
| Rv0881 putative rRNA methylase | Rv1817 fumarate reductase/succinate dehydrogenase flavoprotein, N-terminal:FAD dependent oxidoreductase |
| Rv2689c RNA methyltransferase, TrmA family | Rv0226c Possible membrane protein |
| Rv0227c PROBABLE CONSERVED MEMBRANE PROTEIN | Rv0879c POSSIBLE CONSERVED TRANSMEMBRANE PROTEIN |
| Rv2343c DNA primase (EC 2.7.7.-) | Rv2344c dNTP triphosphohydrolase, broad substrate specificity, subgroup 2 |
| Rv2696c | Rv2761c Type I restriction-modification system, specificity subunit S (EC 3.1.21.3) |
| Rv2762c |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006304 | DNA modification |
|
GO:0040007 | growth |
|
GO:0005886 | plasma membrane |
|
GO:0047570 | 3-oxoadipate enol-lactonase activity |
|
GO:0005618 | cell wall |
|
GO:0005887 | integral to plasma membrane |
|
GO:0006260 | DNA replication |
|
GO:0008270 | zinc ion binding |
|
GO:0003676 | nucleic acid binding |
|
GO:0003677 | DNA binding |
|
GO:0006304 | DNA modification |
|
GO:0003896 | DNA primase activity |
|
GO:0008832 | dGTPase activity |
|
GO:0005829 | cytosol |

















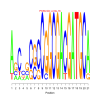
Discussion