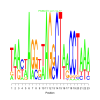
Module 267 Residual: 0.52
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0267 | v02 | 0.52 | -18.75 |
| Rv0846c Multicopper oxidase | Rv1342c CONSERVED MEMBRANE PROTEIN |
| Rv1343c Lipoprotein LprD | Rv1348 ABC transporter ATP-binding protein |
| Rv1349 ABC transporter ATP-binding protein | Rv1519 |
| Rv2121c ATP phosphoribosyltransferase (EC 2.4.2.17) | Rv2122c Phosphoribosyl-ATP pyrophosphatase (EC 3.6.1.31) |
| Rv2123 PPE family protein | Rv3402c DegT/DnrJ/EryC1 family protein |
| Rv3752c tRNA-specific adenosine-34 deaminase (EC 3.5.4.-) | Rv3403c Zn-dependent hydrolases, including glyoxylases |
| Rv3754 Arogenate dehydrogenase (EC 1.3.1.43) | Rv3837c putative phosphoglycerate mutase |
| Rv3838c Prephenate dehydratase (EC 4.2.1.51) | Rv3839 |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0046394 | GO:0016053 | GO:0000105 | GO:0006547 | GO:0052803 | GO:0006520 | GO:0044283 | GO:0044711 | GO:0009073 | carboxylic acid biosynthetic process | organic acid biosynthetic process | histidine biosynthetic process | histidine metabolic process | imidazole-containing compound metabolic ... | cellular amino acid metabolic process | small molecule biosynthetic process | single-organism biosynthetic process | aromatic amino acid family biosynthetic ... |
|
GO:0016491 | oxidoreductase activity |
|
GO:0005507 | copper ion binding |
|
GO:0005887 | integral to plasma membrane |
|
GO:0040007 | growth |
|
GO:0005886 | plasma membrane |
|
GO:0009405 | pathogenesis |
|
GO:0010106 | cellular response to iron ion starvation |
|
GO:0015891 | siderophore transport |
|
GO:0042927 | siderophore transporter activity |
|
GO:0052099 | acquisition by symbiont of nutrients from host via siderophores |
|
GO:0055072 | iron ion homeostasis |
|
GO:0075139 | response to host iron concentration |
|
GO:0005829 | cytosol |
|
GO:0003879 | ATP phosphoribosyltransferase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0005524 | ATP binding |
|
GO:0016208 | AMP binding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0051260 | protein homooligomerization |
|
GO:0004636 | phosphoribosyl-ATP diphosphatase activity |
|
GO:0052572 | response to host immune response |
|
GO:0005576 | extracellular region |
|
GO:0008270 | zinc ion binding |
|
GO:0016787 | hydrolase activity |
|
GO:0008977 | prephenate dehydrogenase activity |
|
GO:0004664 | prephenate dehydratase activity |
|
GO:0016597 | amino acid binding |
|
GO:0033585 | L-phenylalanine biosynthetic process from chorismate via phenylpyruvate |
|
GO:0051289 | protein homotetramerization |
|
GO:0004106 | chorismate mutase activity |


















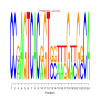
Discussion