Module 351 Residual: 0.53
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0351 | v02 | 0.53 | -19.66 |
| Rv1003 rRNA small subunit methyltransferase I | Rv3432c Glutamate decarboxylase (EC 4.1.1.15) |
| Rv3813c Cof family hydrolase | Rv3815c 1-acyl-sn-glycerol-3-phosphate acyltransferase (EC 2.3.1.51) |
| Rv3817 aminoglycoside 3'-phosphotransferase | Rv1914c |
| Rv1915 Isocitrate lyase (EC 4.1.3.1), group III, Mycobacterial type ICL2 | Rv1916 Isocitrate lyase (EC 4.1.3.1), group III, Mycobacterial type ICL2 |
| Rv2298 Aldo/keto reductase | Rv2609c FIG019327: Probable conserved membrane protein |
| Rv2610c Phosphatidylinositol alpha-mannosyltransferase (EC 2.4.1.57) | Rv2611c Lauroyl/myristoyl acyltransferase involved in lipid A biosynthesis (Lauroyl/myristoyl acyltransferase) |
| Rv2613c FIG049476: HIT family protein | Rv2614c Threonyl-tRNA synthetase (EC 6.1.1.3) |
| Rv2895c Iron utilization protein | Rv3814c 1-acyl-sn-glycerol-3-phosphate acyltransferase (EC 2.3.1.51) |
| Rv3816c 1-acyl-sn-glycerol-3-phosphate acyltransferase (EC 2.3.1.51) |
|
GO:0005886 | plasma membrane |
|
GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity |
|
GO:0004451 | isocitrate lyase activity |
|
GO:0006097 | glyoxylate cycle |
|
GO:0046421 | methylisocitrate lyase activity |
|
GO:0005618 | cell wall |
|
GO:0000030 | mannosyltransferase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0043750 | phosphatidylinositol alpha-mannosyltransferase activity |
|
GO:0040007 | growth |
|
GO:0004829 | threonine-tRNA ligase activity |
|
GO:0010106 | cellular response to iron ion starvation |
|
GO:0015891 | siderophore transport |



















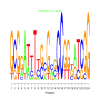
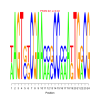
Discussion