Module 370 Residual: 0.49
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0370 | v02 | 0.49 | -24.10 |
| Rv1459c POSSIBLE CONSERVED INTEGRAL MEMBRANE PROTEIN | Rv1461 Iron-sulfur cluster assembly protein SufB @ intein-containing |
| Rv1462 Iron-sulfur cluster assembly protein SufD | Rv1465 Putative iron-sulfur cluster assembly scaffold protein for SUF system, SufE2 |
| Rv1466 PaaD-like protein (DUF59) involved in Fe-S cluster assembly | Rv2531c Arginine decarboxylase (EC 4.1.1.19) |
| Rv3600c Pantothenate kinase type III, CoaX-like (EC 2.7.1.33) | Rv3601c Aspartate 1-decarboxylase (EC 4.1.1.11) |
| Rv3602c Pantoate--beta-alanine ligase (EC 6.3.2.1) | Rv3603c |
| Rv0483 Possible lipoprotein LprQ | Rv1464 Cysteine desulfurase (EC 2.8.1.7), SufS subfamily |
| Rv1778c | Rv3599c |
|
GO:0040007 | growth |
|
GO:0004519 | endonuclease activity |
|
GO:0005515 | protein binding |
|
GO:0005829 | cytosol |
|
GO:0042803 | protein homodimerization activity |
|
GO:0003674 | molecular_function |
|
GO:0052572 | response to host immune response |
|
GO:0005886 | plasma membrane |
|
GO:0004594 | pantothenate kinase activity |
|
GO:0004068 | aspartate 1-decarboxylase activity |
|
GO:0005618 | cell wall |
|
GO:0051289 | protein homotetramerization |
|
GO:0004592 | pantoate-beta-alanine ligase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0005524 | ATP binding |
|
GO:0015940 | pantothenate biosynthetic process |
|
GO:0019482 | beta-alanine metabolic process |
|
GO:0030145 | manganese ion binding |
|
GO:0031071 | cysteine desulfurase activity |
















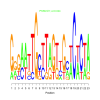
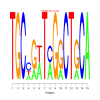
Discussion