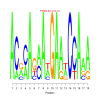
Module 413 Residual: 0.50
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0413 | v02 | 0.50 | -22.76 |
| Rv1230c POSSIBLE MEMBRANE PROTEIN | Rv3755c |
| Rv3756c Glycine betaine ABC transport system permease protein | Rv3759c L-proline glycine betaine binding ABC transporter protein ProX (TC 3.A.1.12.1) |
| Rv1891 Possible membrane protein | Rv1892 Possible membrane protein |
| Rv1893 | Rv3083 Monooxygenase, flavin-binding family |
| Rv3084 Esterase/lipase | Rv3085 PROBABLE SHORT-CHAIN TYPE DEHYDROGENASE/REDUCTASE (EC 1.-.-.-) |
| Rv3086 Alcohol dehydrogenase (EC 1.1.1.1) | Rv3087 Wax ester synthase/acyl-CoA:diacylglycerol acyltransferase |
| Rv3088 Wax ester synthase/acyl-CoA:diacylglycerol acyltransferase | Rv3089 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) |
| Rv3758c L-proline glycine betaine ABC transport system permease protein ProV (TC 3.A.1.12.1) | Rv3760 Cell division protein DivIC (FtsB), stabilizes FtsL against RasP cleavage |
|
GO:0031460 | glycine betaine transport |
|
GO:0075136 | response to host |
|
GO:0005576 | extracellular region |
|
GO:0009405 | pathogenesis |
|
GO:0001101 | response to acid |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0005886 | plasma membrane |
|
GO:0004144 | diacylglycerol O-acyltransferase activity |
|
GO:0045017 | glycerolipid biosynthetic process |
|
GO:0071731 | response to nitric oxide |
|
GO:0004467 | long-chain fatty acid-CoA ligase activity |
|
GO:0005829 | cytosol |
|
GO:0005524 | ATP binding |
|
GO:0016887 | ATPase activity |


















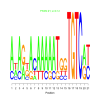
Discussion