Module 472 Residual: 0.59
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0472 | v02 | 0.59 | -10.03 |
| Rv1747 Probable conserved transmembrane ATP-binding protein ABC transporter | Rv3875 6 kDa early secretory antigenic target ESAT-6 (EsxA) |
| Rv3876 RD1 region associated protein Rv3876 | Rv1248c Dihydrolipoamide succinyltransferase component (E2) of 2-oxoglutarate dehydrogenase complex (EC 2.3.1.61) / 2-oxoglutarate dehydrogenase E1 component (EC 1.2.4.2) |
| Rv1333 possible hydrolase | Rv1335 9.5 kDa culture filtrate antigen Cfp10A |
| Rv2081c | Rv2082 |
| Rv2083 | Rv2163c Cell division protein FtsI [Peptidoglycan synthetase] (EC 2.4.1.129) |
| Rv2164c FIG034299: proline rich membrane protein | Rv2345 Possible membrane protein |
| Rv2365c | Rv3676 cAMP-binding proteins - catabolite gene activator and regulatory subunit of cAMP-dependent protein kinases |
| Rv3719 FAD/FMN-containing dehydrogenases | Rv3720 Cyclopropane-fatty-acyl-phospholipid synthase (EC 2.1.1.79) |
| Rv3877 Putative ESX-1 secretion system component Rv3877 | Rv3884c Cell division protein FtsH (EC 3.4.24.-) |
| Rv3910 Proposed peptidoglycan lipid II flippase MurJ |
|
GO:0005515 | protein binding |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0005576 | extracellular region |
|
GO:0005737 | cytoplasm |
|
GO:0005886 | plasma membrane |
|
GO:0009405 | pathogenesis |
|
GO:0042803 | protein homodimerization activity |
|
GO:0046812 | host cell surface binding |
|
GO:0052027 | modulation by symbiont of host signal transduction pathway |
|
GO:0052083 | negative regulation by symbiont of host cell-mediated immune response |
|
GO:0000287 | magnesium ion binding |
|
GO:0006099 | tricarboxylic acid cycle |
|
GO:0008683 | 2-oxoglutarate decarboxylase activity |
|
GO:0009055 | electron carrier activity |
|
GO:0040007 | growth |
|
GO:0045254 | pyruvate dehydrogenase complex |
|
GO:0055114 | oxidation-reduction process |
|
GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity |
|
GO:0019344 | cysteine biosynthetic process |
|
GO:0043234 | protein complex |
|
GO:0008955 | peptidoglycan glycosyltransferase activity |
|
GO:0005887 | integral to plasma membrane |
|
GO:0003677 | DNA binding |
|
GO:0045892 | negative regulation of transcription, DNA-dependent |
|
GO:0030552 | cAMP binding |
|
GO:0045893 | positive regulation of transcription, DNA-dependent |
|
GO:0042783 | active evasion of host immune response |
|
GO:0044110 | growth involved in symbiotic interaction |
|
GO:0044117 | growth of symbiont in host |





















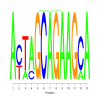
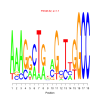
Discussion