Module 511 Residual: 0.52
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0511 | v02 | 0.52 | -9.33 |
| Rv1811 Mg(2+) transport ATPase protein C | Rv0042c Transcriptional regulator, MarR family |
| Rv0089 Methyltransferase/methylase | Rv0318c Possible membrane protein |
| Rv0347 | Rv0349 |
| Rv0381c | Rv0979c |
| Rv0999 | Rv1019 Transcriptional regulator, TetR family |
| Rv1020 Transcription-repair coupling factor | Rv1021 Nucleoside triphosphate pyrophosphohydrolase MazG (EC 3.6.1.8) |
| Rv1136 Enoyl-CoA hydratase (EC 4.2.1.17) | Rv1353c Transcriptional regulator, TetR family |
| Rv1356c | Rv3731 ATP-dependent DNA ligase (EC 6.5.1.1) LigC |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0031177 | GO:0072341 | GO:0033218 | GO:0016597 | phosphopantetheine binding | modified amino acid binding | amide binding | amino acid binding |
|
GO:0009405 | pathogenesis |
|
GO:0010350 | cellular response to magnesium starvation |
|
GO:0005886 | plasma membrane |
|
GO:0005618 | cell wall |
|
GO:0040007 | growth |
|
GO:0005576 | extracellular region |
|
GO:0003700 | sequence-specific DNA binding transcription factor activity |
|
GO:0008026 | ATP-dependent helicase activity |
|
GO:0003676 | nucleic acid binding |
|
GO:0004386 | helicase activity |
|
GO:0005524 | ATP binding |
|
GO:0005829 | cytosol |


















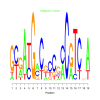
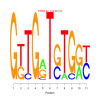
Discussion