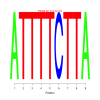
Module 551 Residual: 0.63
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0551 | v02 | 0.63 | -6.37 |
| Rv0198c Metallopeptidase | Rv0633c Possible membrane protein |
| Rv0993 UTP--glucose-1-phosphate uridylyltransferase (EC 2.7.7.9) | Rv1271c |
| Rv2190c Probable endopeptidase | Rv2427c Gamma-glutamyl phosphate reductase (EC 1.2.1.41) |
| Rv2477c ABC transporter ATP-binding protein | Rv2536 Possible membrane protein |
| Rv2982c Glycerol-3-phosphate dehydrogenase [NAD(P)+] (EC 1.1.1.94) | Rv3116 Sulfur carrier protein adenylyltransferase ThiF |
| Rv3274c Butyryl-CoA dehydrogenase (EC 1.3.99.2) | Rv3285 Biotin carboxylase of acetyl-CoA carboxylase (EC 6.3.4.14) / Biotin carboxyl carrier protein of acetyl-CoA carboxylase |
| Rv3311 | Rv3389c Enoyl-CoA hydratase |
| Rv3682 Multimodular transpeptidase-transglycosylase (EC 2.4.1.129) (EC 3.4.-.-) | Rv3850 |
|
GO:0005618 | cell wall |
|
GO:0005886 | plasma membrane |
|
GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity |
|
GO:0040007 | growth |
|
GO:0005576 | extracellular region |
|
GO:0004350 | glutamate-5-semialdehyde dehydrogenase activity |
|
GO:0005829 | cytosol |
|
GO:0005887 | integral to plasma membrane |
|
GO:0046812 | host cell surface binding |
|
GO:0047952 | glycerol-3-phosphate dehydrogenase [NAD(P)+] activity |
|
GO:0003824 | catalytic activity |
|
GO:0004085 | butyryl-CoA dehydrogenase activity |
|
GO:0003989 | acetyl-CoA carboxylase activity |
|
GO:0005515 | protein binding |
|
GO:0009317 | acetyl-CoA carboxylase complex |
|
GO:0006633 | fatty acid biosynthetic process |
|
GO:0016508 | long-chain-enoyl-CoA hydratase activity |
|
GO:0019171 | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase activity |
|
GO:0008955 | peptidoglycan glycosyltransferase activity |


















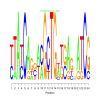
Discussion