Module 581 Residual: 0.61
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0581 | v02 | 0.61 | -11.06 |
| Rv2326c Candidate substrate-specific domain of ECF transporters in Mycobacteria / Duplicated ATPase component of energizing module of predicted ECF transporter in Mycobacteria | Rv2916c Signal recognition particle, subunit Ffh SRP54 (TC 3.A.5.1.1) |
| Rv0204c Possible membrane protein | Rv0505c Phosphoserine phosphatase (EC 3.1.3.3) |
| Rv0509 Glutamyl-tRNA reductase (EC 1.2.1.70) | Rv0510 Porphobilinogen deaminase (EC 2.5.1.61) |
| Rv0511 Uroporphyrinogen-III methyltransferase (EC 2.1.1.107) / Uroporphyrinogen-III synthase (EC 4.2.1.75) | Rv0512 Porphobilinogen synthase (EC 4.2.1.24) |
| Rv0513 Transmembrane transport protein MmpL2 | Rv0514 Possible membrane protein |
| Rv0994 Molybdopterin biosynthesis protein MoeA | Rv1093 Serine hydroxymethyltransferase 1 (EC 2.1.2.1) |
| Rv2697c Deoxyuridine 5'-triphosphate nucleotidohydrolase (EC 3.6.1.23) | Rv2698 PROBABLE CONSERVED ALANINE RICH TRANSMEMBRANE PROTEIN |
| Rv2781c 2-nitropropane dioxygenase (EC 1.13.11.32) | Rv3275c Phosphoribosylaminoimidazole carboxylase catalytic subunit (EC 4.1.1.21) |
| Rv3276c Phosphoribosylaminoimidazole carboxylase ATPase subunit (EC 4.1.1.21) | Rv3328c putative RNA polymerase sigma factor |
| Rv3835 PROBABLE CONSERVED MEMBRANE PROTEIN |
|
GO:0005524 | ATP binding |
|
GO:0016887 | ATPase activity |
|
GO:0005886 | plasma membrane |
|
GO:0005887 | integral to plasma membrane |
|
GO:0003723 | RNA binding |
|
GO:0006605 | protein targeting |
|
GO:0016020 | membrane |
|
GO:0006614 | SRP-dependent cotranslational protein targeting to membrane |
|
GO:0005525 | GTP binding |
|
GO:0005618 | cell wall |
|
GO:0040007 | growth |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0008883 | glutamyl-tRNA reductase activity |
|
GO:0004418 | hydroxymethylbilane synthase activity |
|
GO:0004655 | porphobilinogen synthase activity |
|
GO:0005829 | cytosol |
|
GO:0004372 | glycine hydroxymethyltransferase activity |
|
GO:0006544 | glycine metabolic process |
|
GO:0006563 | L-serine metabolic process |
|
GO:0030170 | pyridoxal phosphate binding |
|
GO:0042783 | active evasion of host immune response |
|
GO:0042803 | protein homodimerization activity |
|
GO:0004170 | dUTP diphosphatase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0046080 | dUTP metabolic process |
|
GO:0004638 | phosphoribosylaminoimidazole carboxylase activity |
|
GO:0042542 | response to hydrogen peroxide |
|
GO:0005576 | extracellular region |





















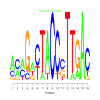
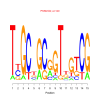
Discussion