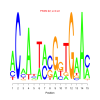
Module 582 Residual: 0.57
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0582 | v02 | 0.57 | -11.33 |
| Rv0423c Hydroxymethylpyrimidine phosphate synthase ThiC | Rv0490 Phosphate regulon sensor protein PhoR (SphS) (EC 2.7.13.3); Sensor-like histidine kinase senX3 (EC 2.7.13.3) |
| Rv1410c EmrB/QacA family drug resistance transporter | Rv1411c Lipoprotein LprG |
| Rv1412 Riboflavin synthase eubacterial/eukaryotic (EC 2.5.1.9) | Rv1487 Putative activity regulator of membrane protease YbbK |
| Rv1488 Putative stomatin/prohibitin-family membrane protease subunit YbbK | Rv1795 Type VII secretion integral membrane protein EccD |
| Rv1797 Type VII secretion protein EccE | Rv1798 Type VII secretion AAA-ATPase EccA |
| Rv1843c Inosine-5'-monophosphate dehydrogenase (EC 1.1.1.205) | Rv2202c Adenosine kinase (EC 2.7.1.20) |
| Rv2535c Aminopeptidase YpdF (MP-, MA-, MS-, AP-, NP- specific) | Rv2868c 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase (EC 1.17.7.1) |
| Rv2941 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) | Rv2942 Putative membrane protein MmpL7 |
| Rv3596c ATP-dependent Clp protease, ATP-binding subunit ClpC / Negative regulator of genetic competence clcC/mecB | Rv3813c Cof family hydrolase |
| Rv3815c 1-acyl-sn-glycerol-3-phosphate acyltransferase (EC 2.3.1.51) |
|
GO:0009228 | thiamine biosynthetic process |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0008150 | biological_process |
|
GO:0005576 | extracellular region |
|
GO:0009405 | pathogenesis |
|
GO:0018106 | peptidyl-histidine phosphorylation |
|
GO:0046777 | protein autophosphorylation |
|
GO:0005887 | integral to plasma membrane |
|
GO:0008144 | drug binding |
|
GO:0015562 | efflux transmembrane transporter activity |
|
GO:0004746 | riboflavin synthase activity |
|
GO:0005618 | cell wall |
|
GO:0003938 | IMP dehydrogenase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0004001 | adenosine kinase activity |
|
GO:0005524 | ATP binding |
|
GO:0005525 | GTP binding |
|
GO:0032567 | dGTP binding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0008235 | metalloexopeptidase activity |
|
GO:0006508 | proteolysis |
|
GO:0046429 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase activity |
|
GO:0004467 | long-chain fatty acid-CoA ligase activity |
|
GO:0004321 | fatty-acyl-CoA synthase activity |
|
GO:0008610 | lipid biosynthetic process |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0052572 | response to host immune response |
|
GO:0070566 | adenylyltransferase activity |
|
GO:0071766 | Actinobacterium-type cell wall biogenesis |
|
GO:0071770 | DIM/DIP cell wall layer assembly |
|
GO:0006855 | drug transmembrane transport |
|
GO:0006869 | lipid transport |
|
GO:0006289 | nucleotide-excision repair |
|
GO:0005515 | protein binding |
|
GO:0004518 | nuclease activity |
|
GO:0019538 | protein metabolic process |
|
GO:0003677 | DNA binding |
|
GO:0016887 | ATPase activity |
|
GO:0044183 | protein binding involved in protein folding |
|
GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity |





















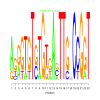
Discussion