Module 25
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.56 | 29 | 65 | NA | NA |
Bicluster expression profile
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
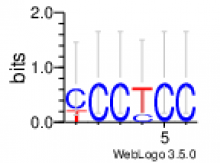
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
The module is significantly enriched with genes associated to the indicated functional terms
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_12480 | rnc | ribonuclease III | Digests double-stranded RNA. Involved in the processing of primary rRNA transcript to yield the immediate precursors to the large and small rRNAs (23S and 16S). Processes some mRNAs, and tRNAs when they are encoded in the rRNA operon. Processes pre-crRNA and tracrRNA of type II CRISPR loci if present in the organism. | Yes |
 |
||
| CD630_31140 | Putative membrane protein |
 |
|||||
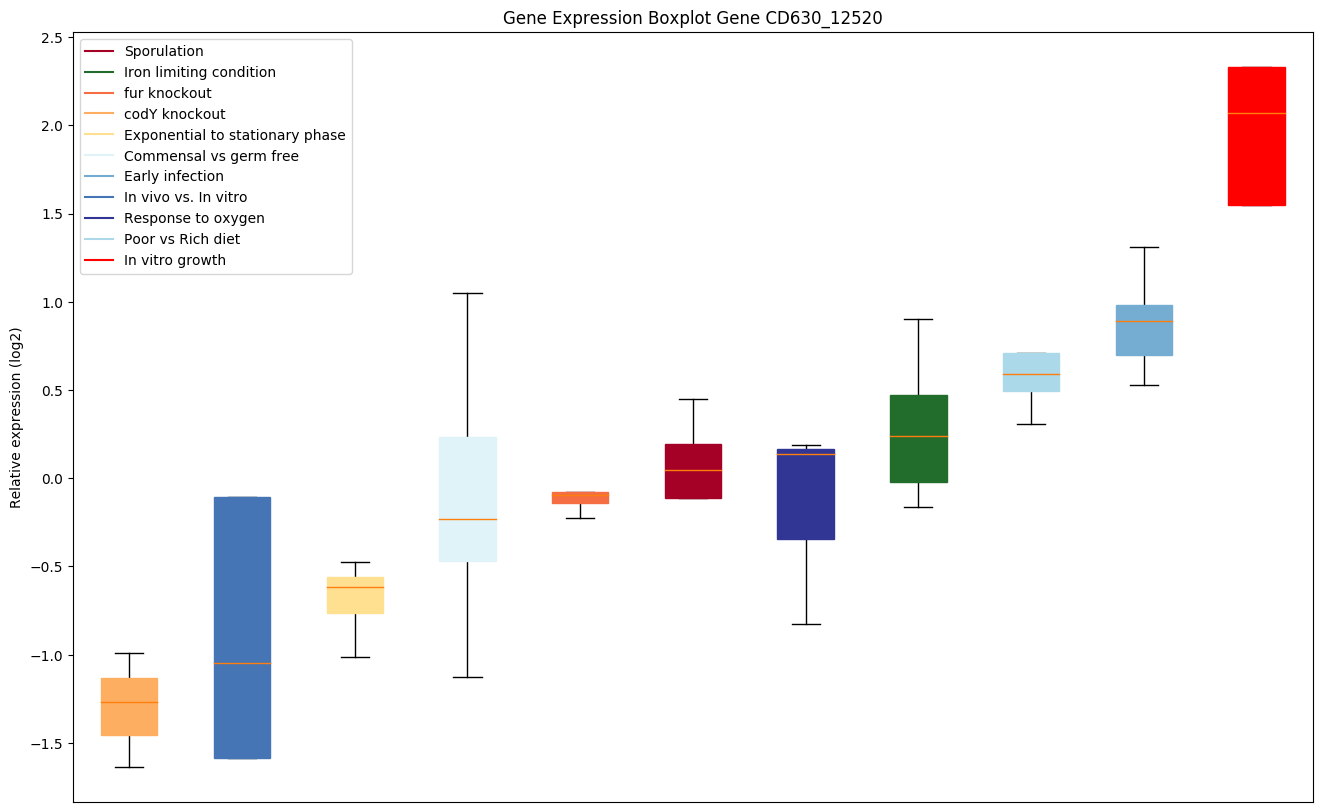
| CD630_12520 | ffh | Signal recognition particle protein | Involved in targeting and insertion of nascent membrane proteins into the cytoplasmic membrane. Binds to the hydrophobic signal sequence of the ribosome-nascent chain (RNC) as it emerges from the ribosomes. The SRP-RNC complex is then targeted to the cytoplasmic membrane where it interacts with the SRP receptor FtsY. | Yes |
 |
||
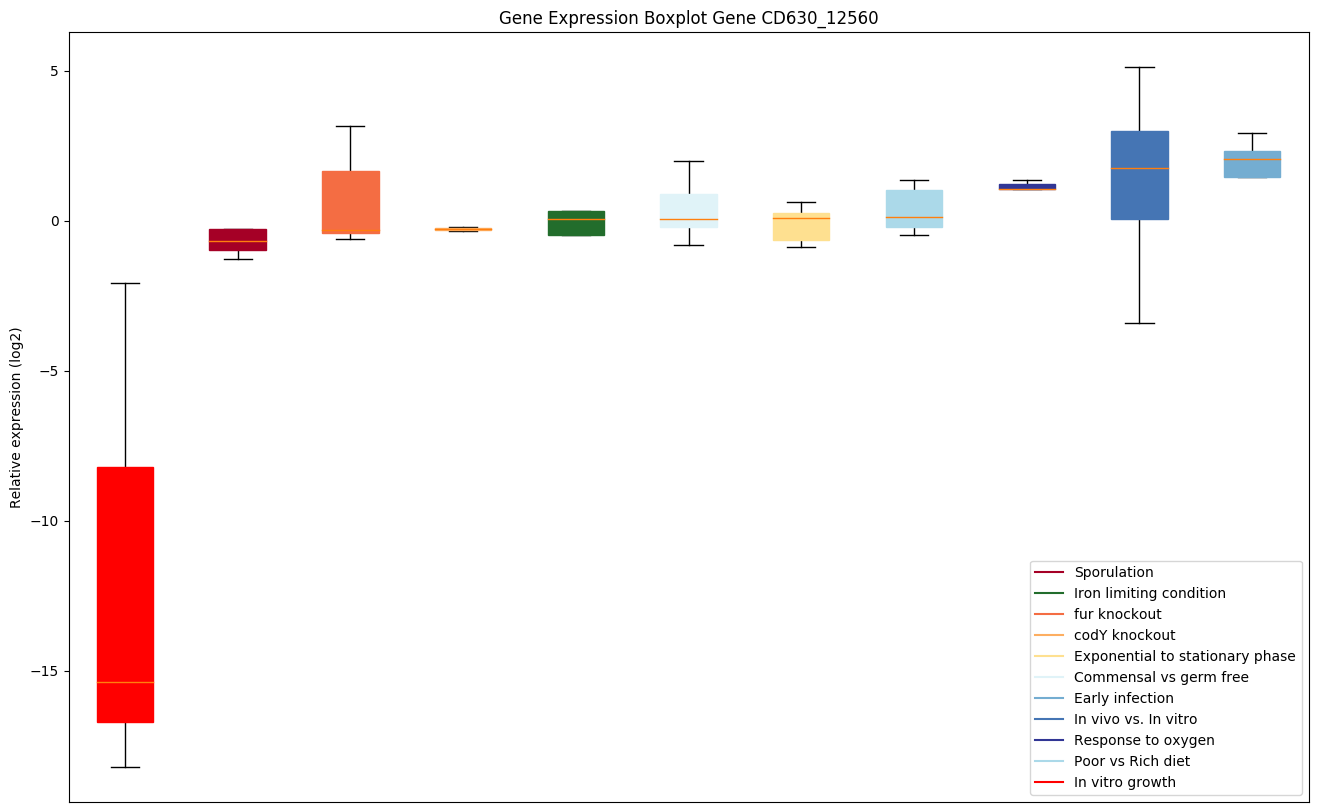
| CD630_12560 | trmD | tRNA (guanine-N(1)-)-methyltransferase(M1G-methyltransferase) (tRNA [GM37] methyltransferase ) | Specifically methylates guanosine-37 in various tRNAs. | Yes |
 |
||
| CD630_12500 | smc | Structural maintenance chromosome protein SMC | Required for chromosome condensation and partitioning. |
 |
|||
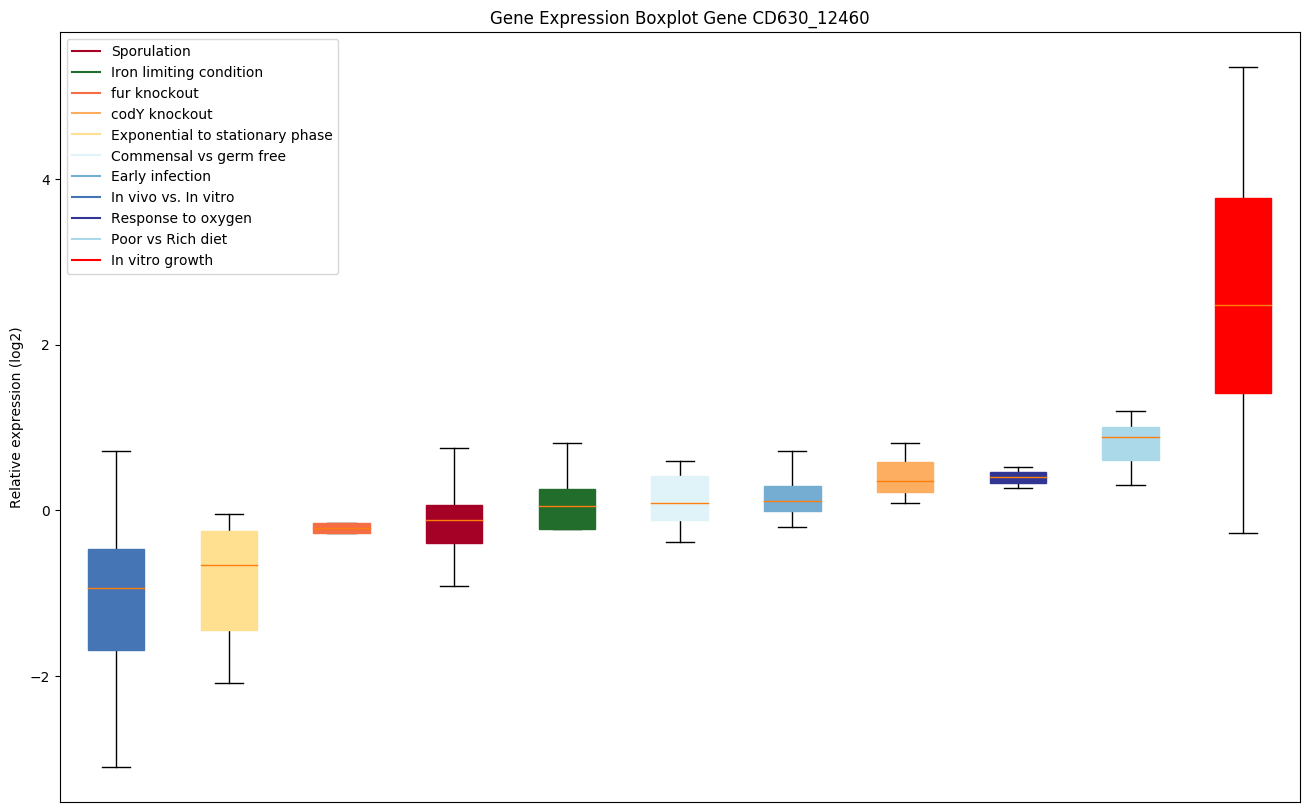
| CD630_12460 | efp | Elongation factor P (EF-P) | Involved in peptide bond synthesis. Stimulates efficient translation and peptide-bond synthesis on native or reconstituted 70S ribosomes in vitro. Probably functions indirectly by altering the affinity of the ribosome for aminoacyl-tRNA, thus increasing their reactivity as acceptors for peptidyl transferase. | Yes |
 |
||
| CD630_12540 | Putative RNA-binding protein |
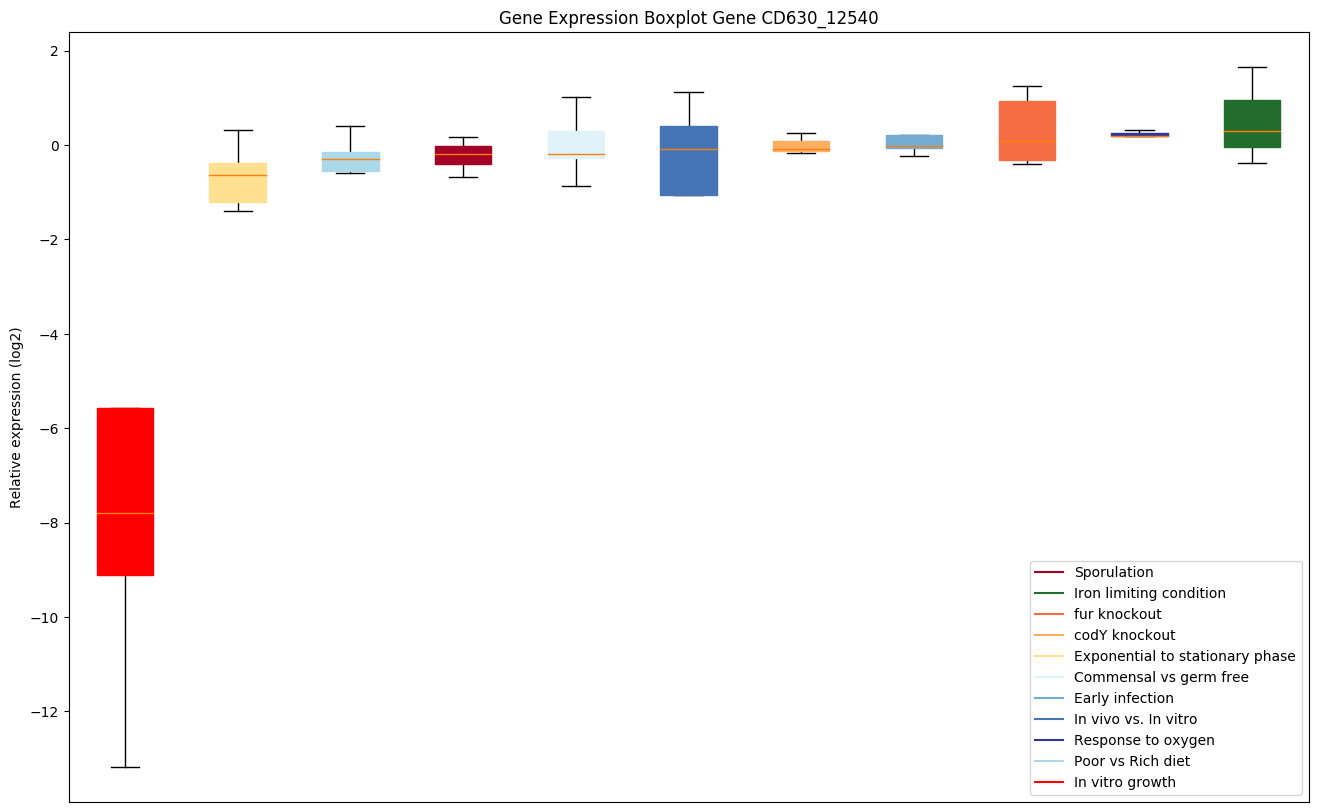 |
|||||
| CD630_31120 | Putative peptidase, M20D family |
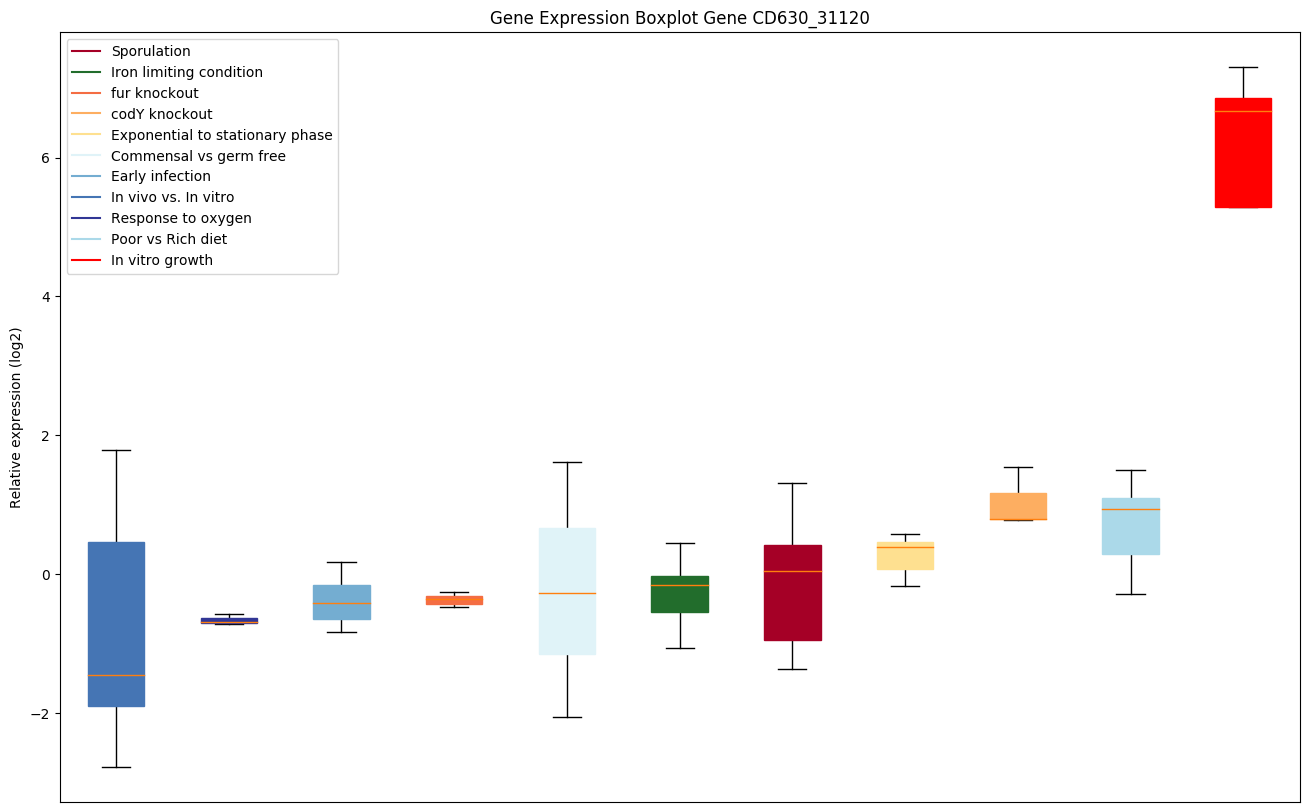 |
|||||
| CD630_31960 | Putative penicillin-binding protein |
 |
|||||
| CD630_20190 | Transporter drug resistance, Major FacilitatorSuperfamily (MFS) |
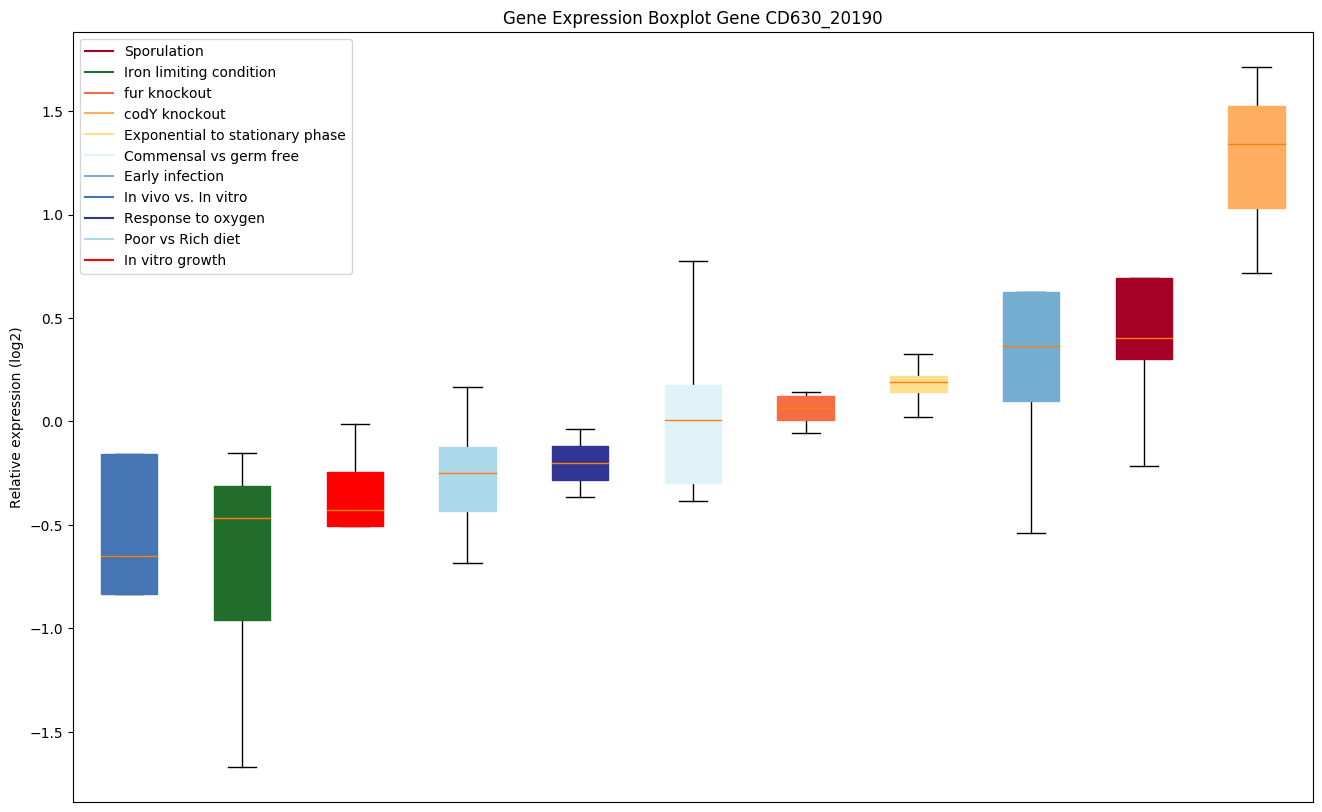 |
|||||
| CD630_31940 | Transcriptional regulator, AraC family |
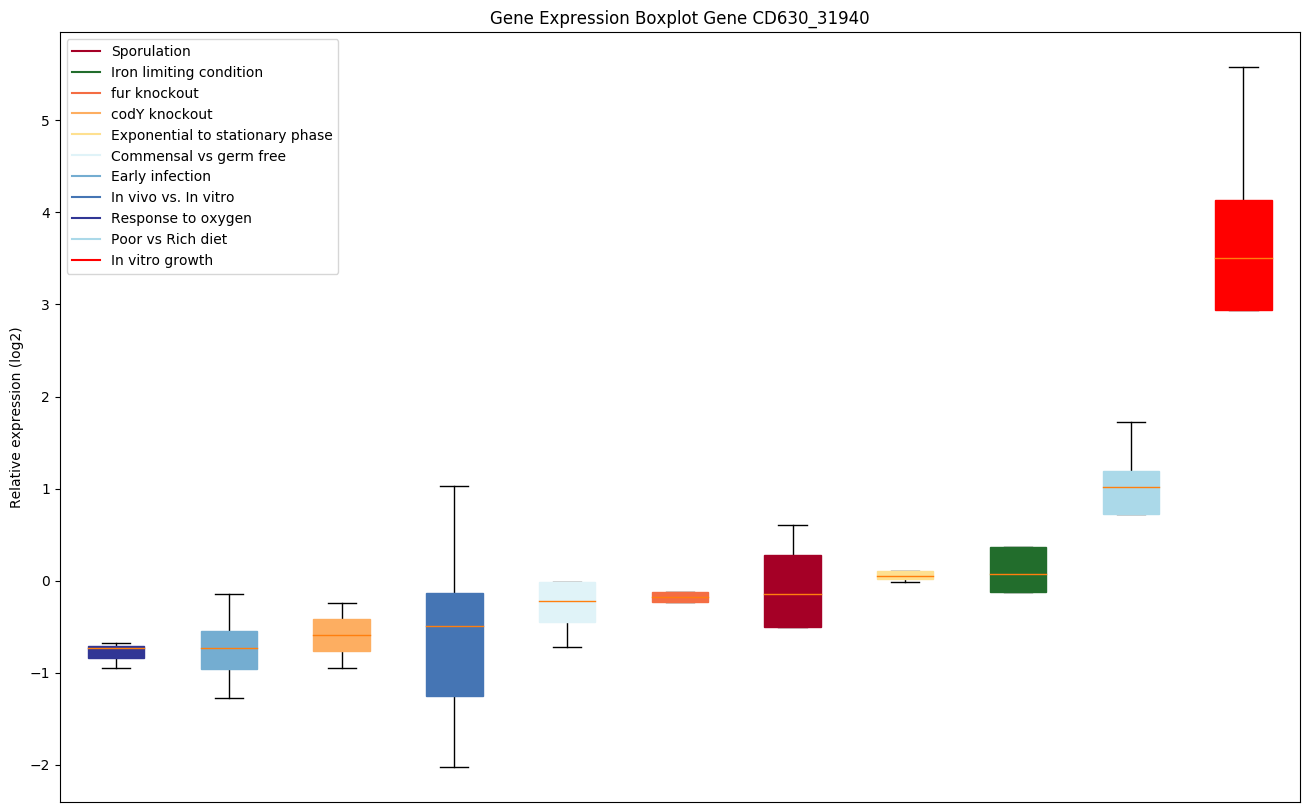 |
|||||
| CD630_08620 | PTS system, lactose/cellobiose-family IICmembrane component | The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. |
 |
||||
| CD630_32620 | pstA | ABC-type transport system, phosphate-specificpermease |
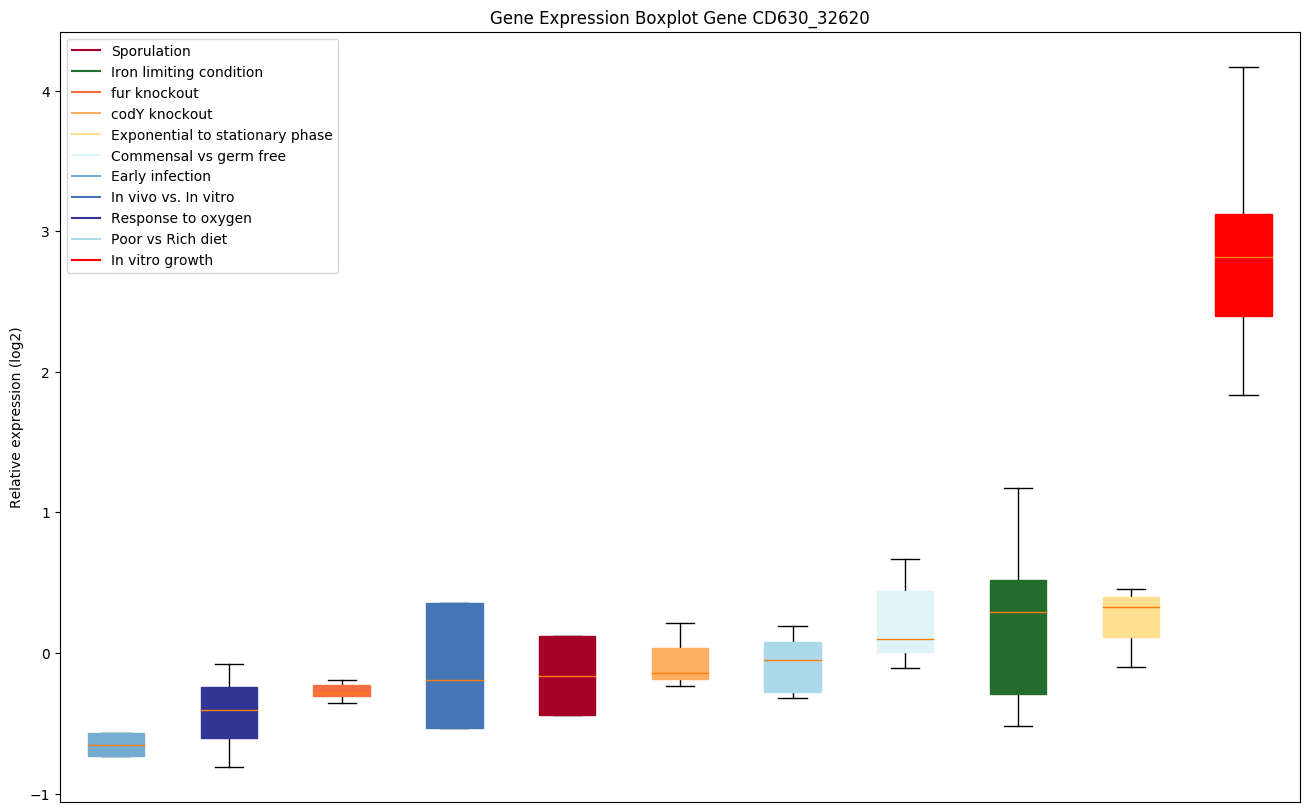 |
||||
| CD630_32600 | phoU | Phosphate uptake regulator, PhoU | Plays a role in the regulation of phosphate uptake. |
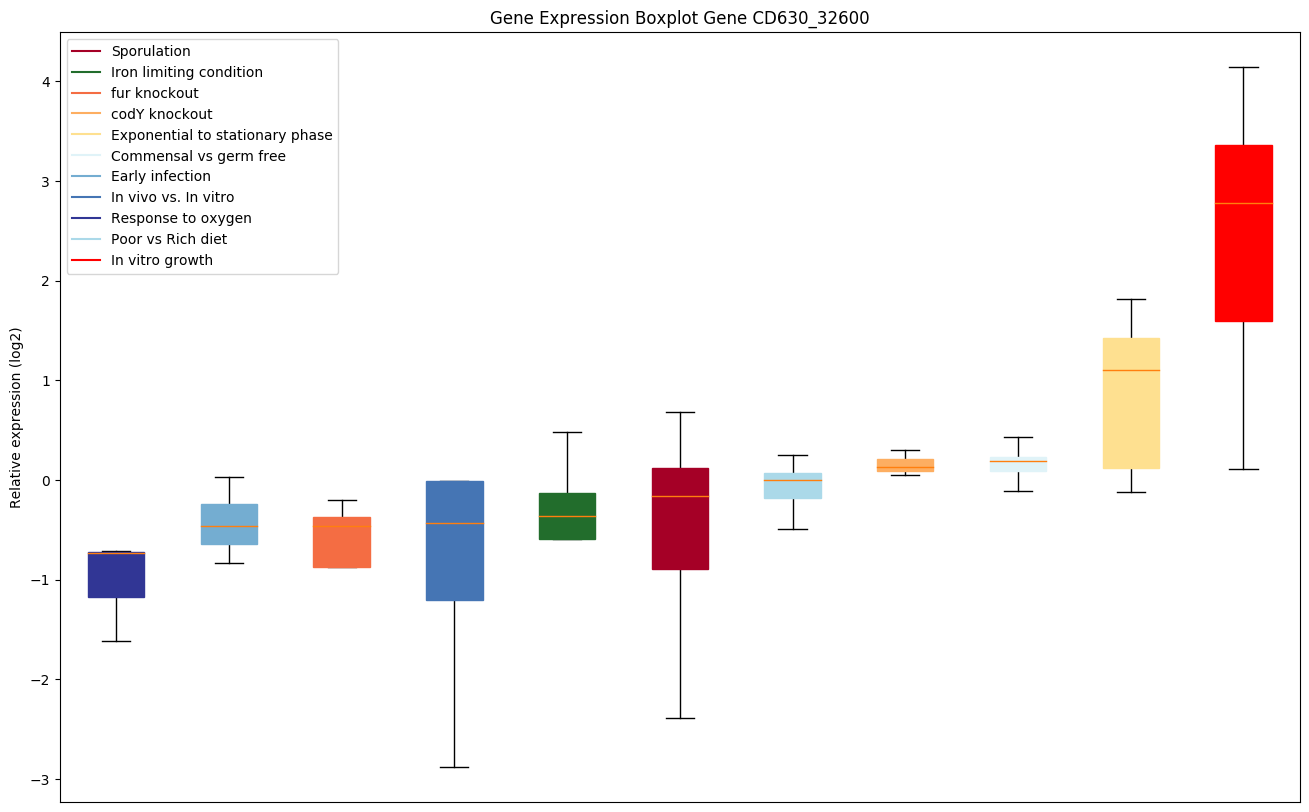 |
|||
| CD630_12530 | rpsP | 30S ribosomal protein S16 | Yes |
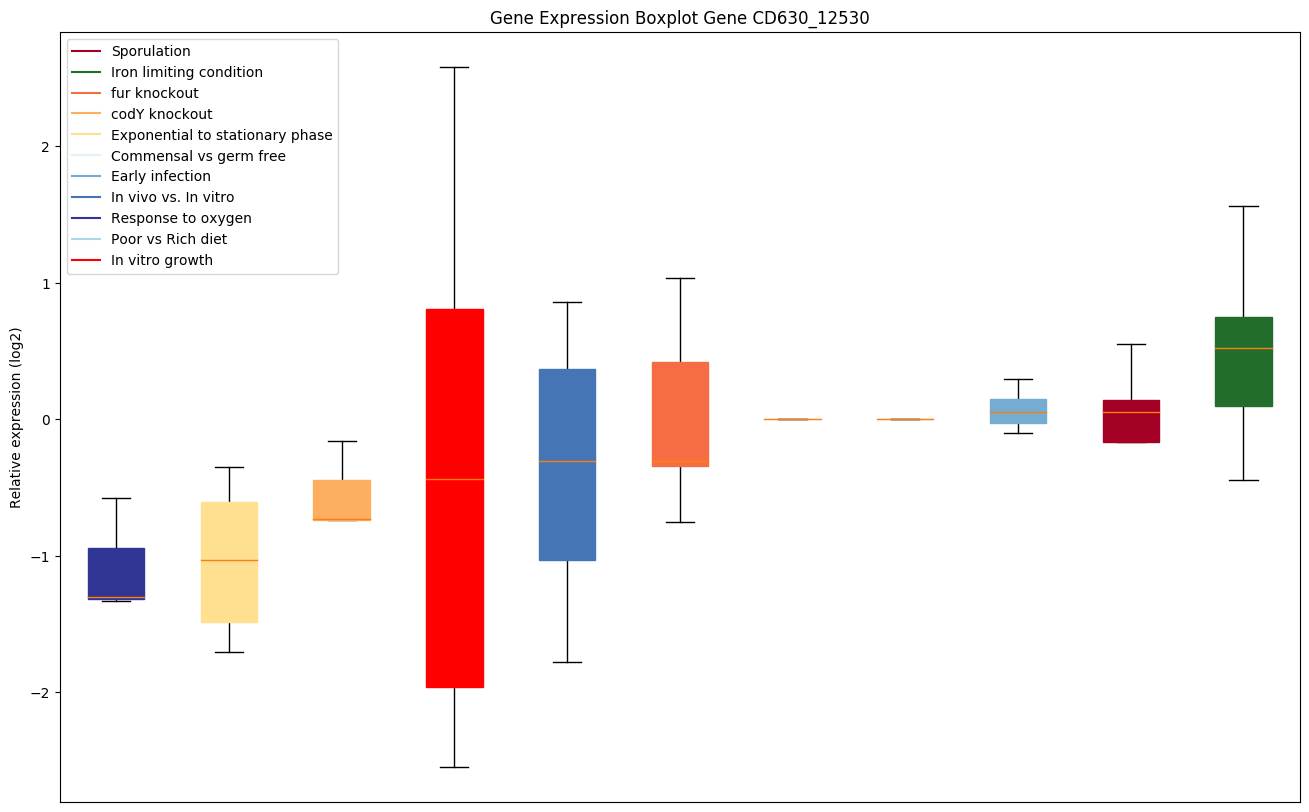 |
|||
| CD630_31980 | cme | Multidrug resistance protein Cme; Transporter,Major Facilitator Superfamily (MFS) |
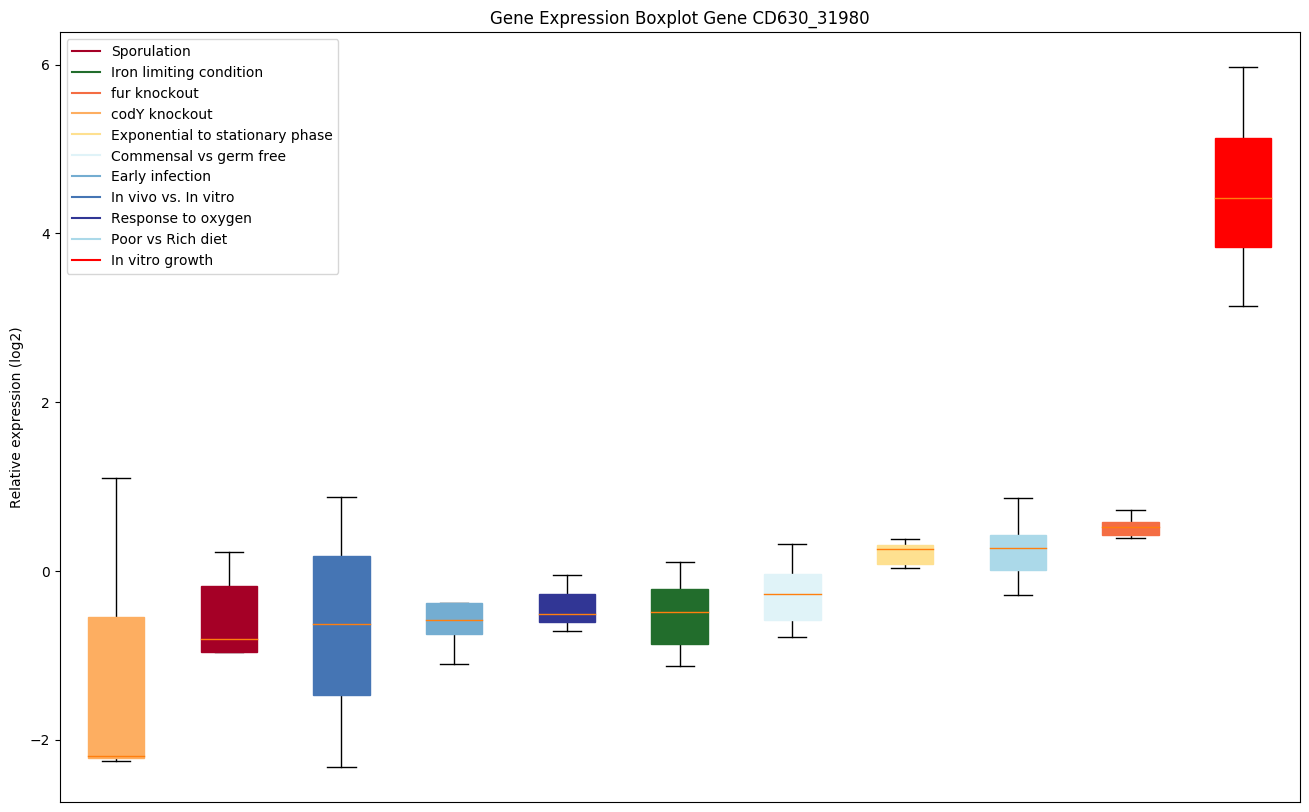 |
||||
| CD630_31130 | Putative aminobenzoyl-glutamate utilizationprotein B |
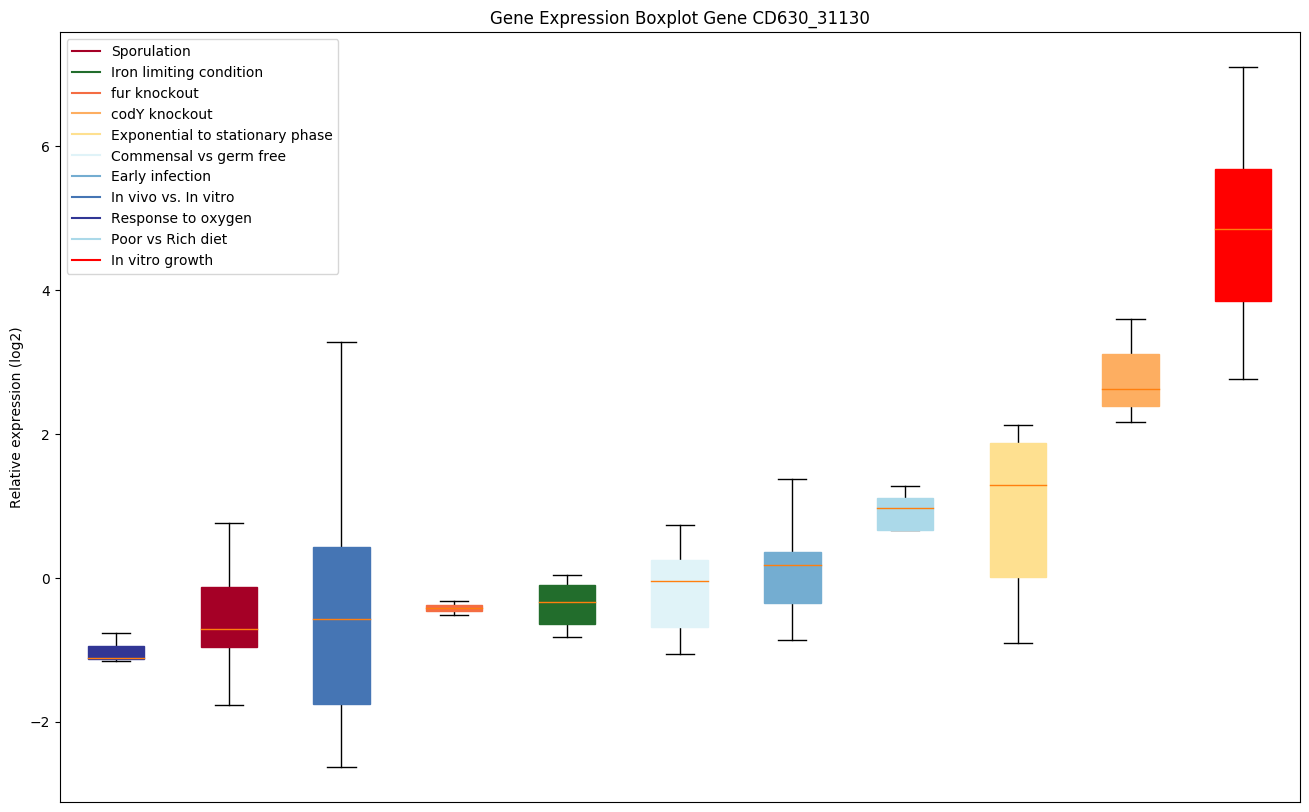 |
|||||
| CD630_12470 | Conserved hypothetical protein |
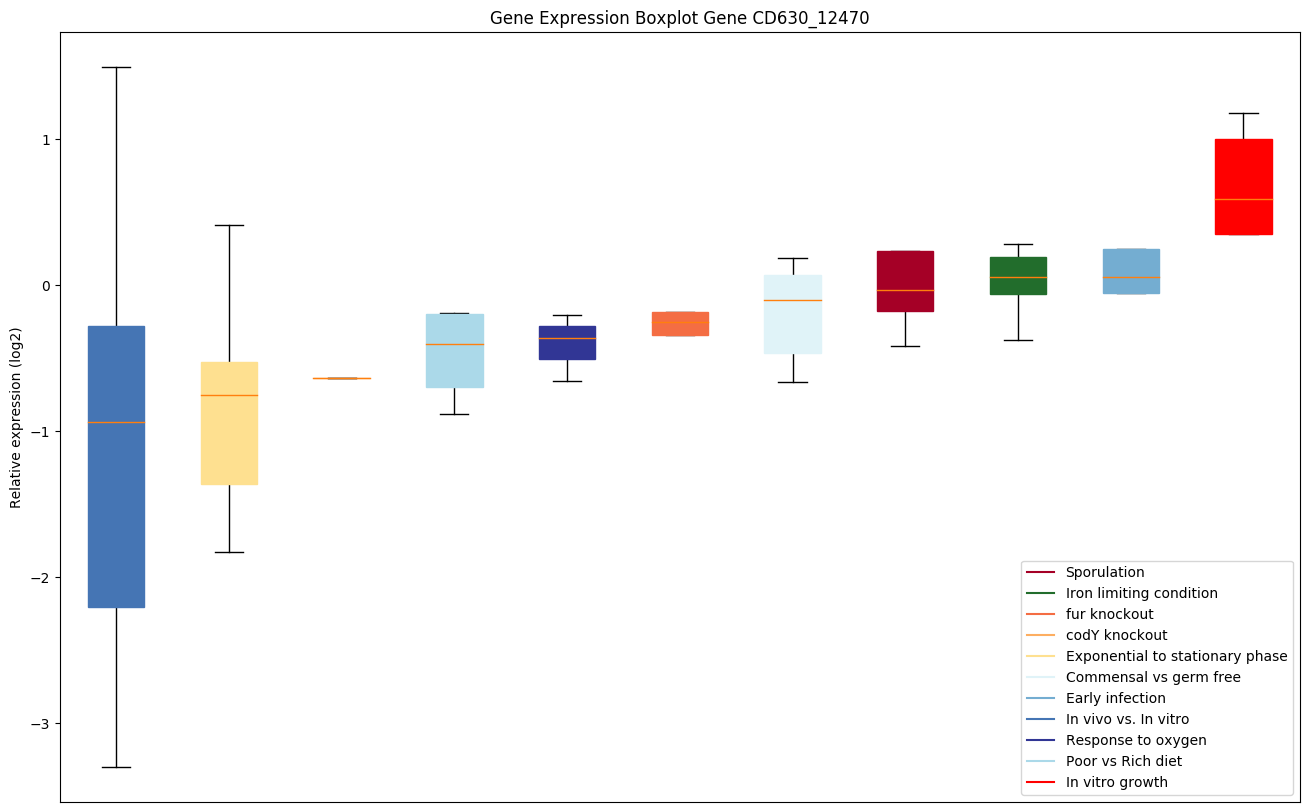 |
|||||
| CD630_12660 | ABC-type transport system, multidrug-familypermease |
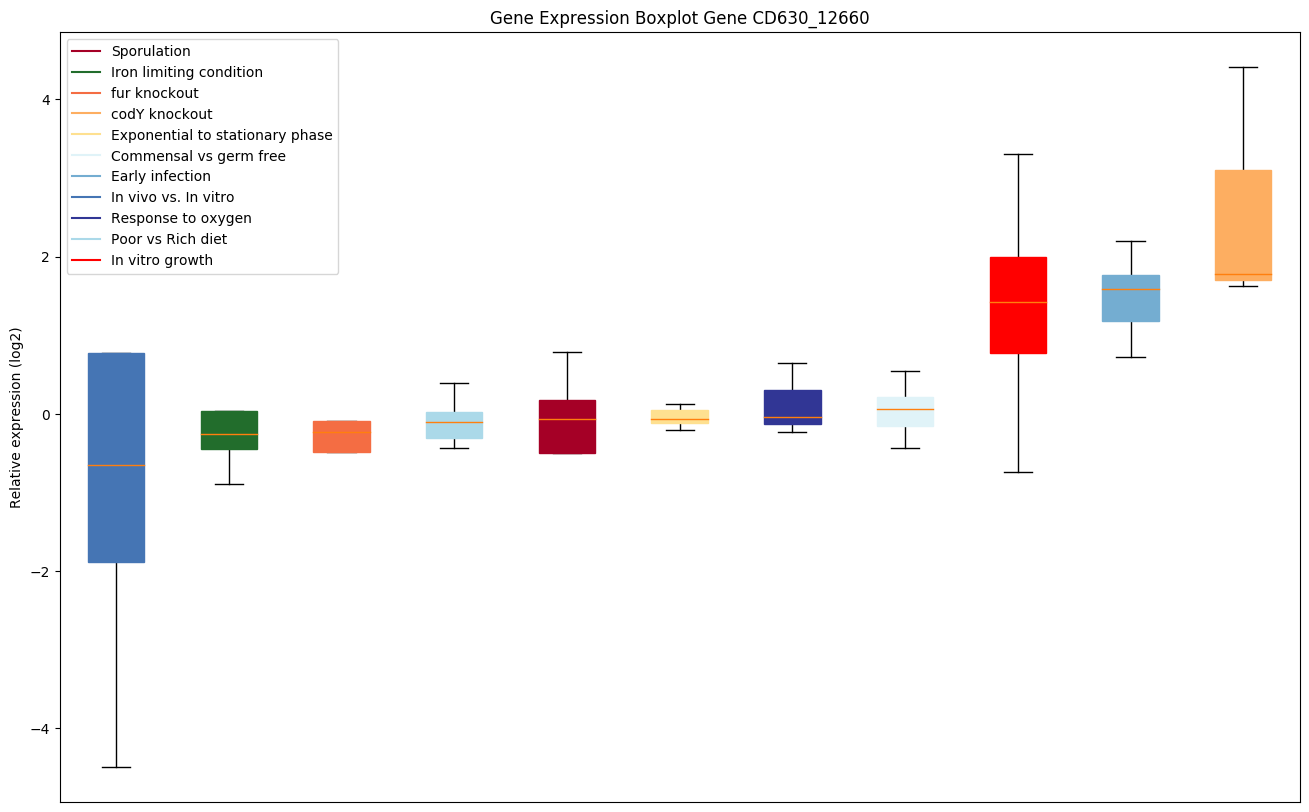 |
|||||
| CD630_12511 | Putative DNA-binding regulator | Might take part in the signal recognition particle (SRP) pathway. This is inferred from the conservation of its genetic proximity to ftsY/ffh. May be a regulatory protein. | Yes |
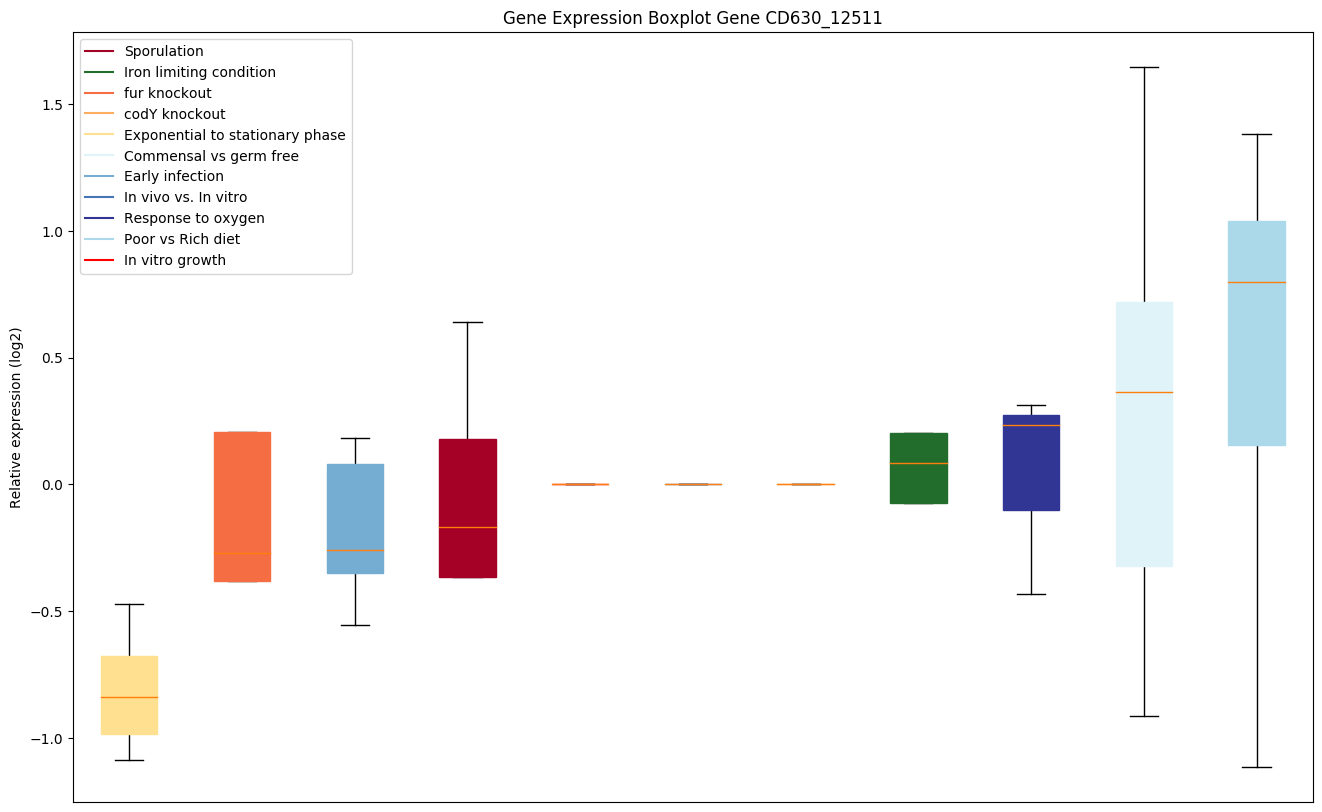 |
|||
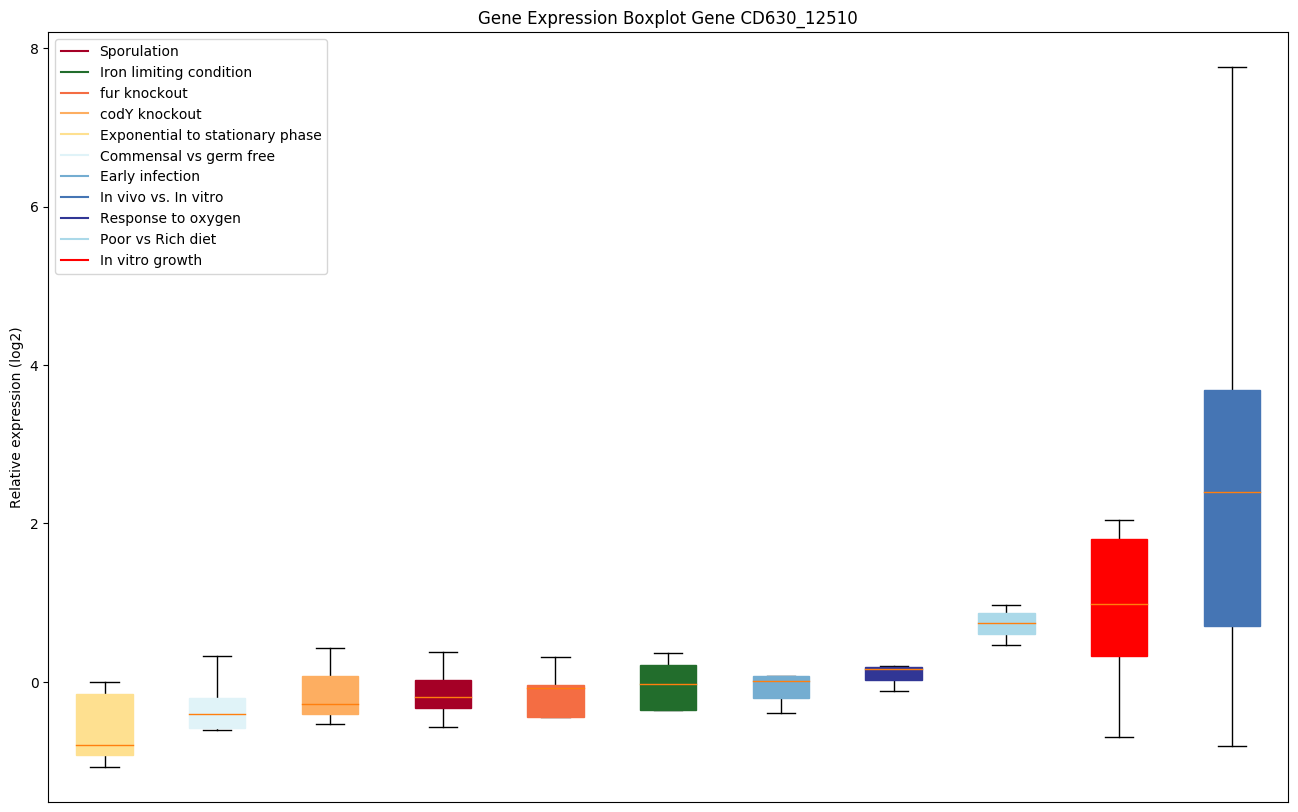
| CD630_12510 | ftsY | signal recognition particle receptor | Involved in targeting and insertion of nascent membrane proteins into the cytoplasmic membrane. Acts as a receptor for the complex formed by the signal recognition particle (SRP) and the ribosome-nascent chain (RNC). | Yes |
 |
||
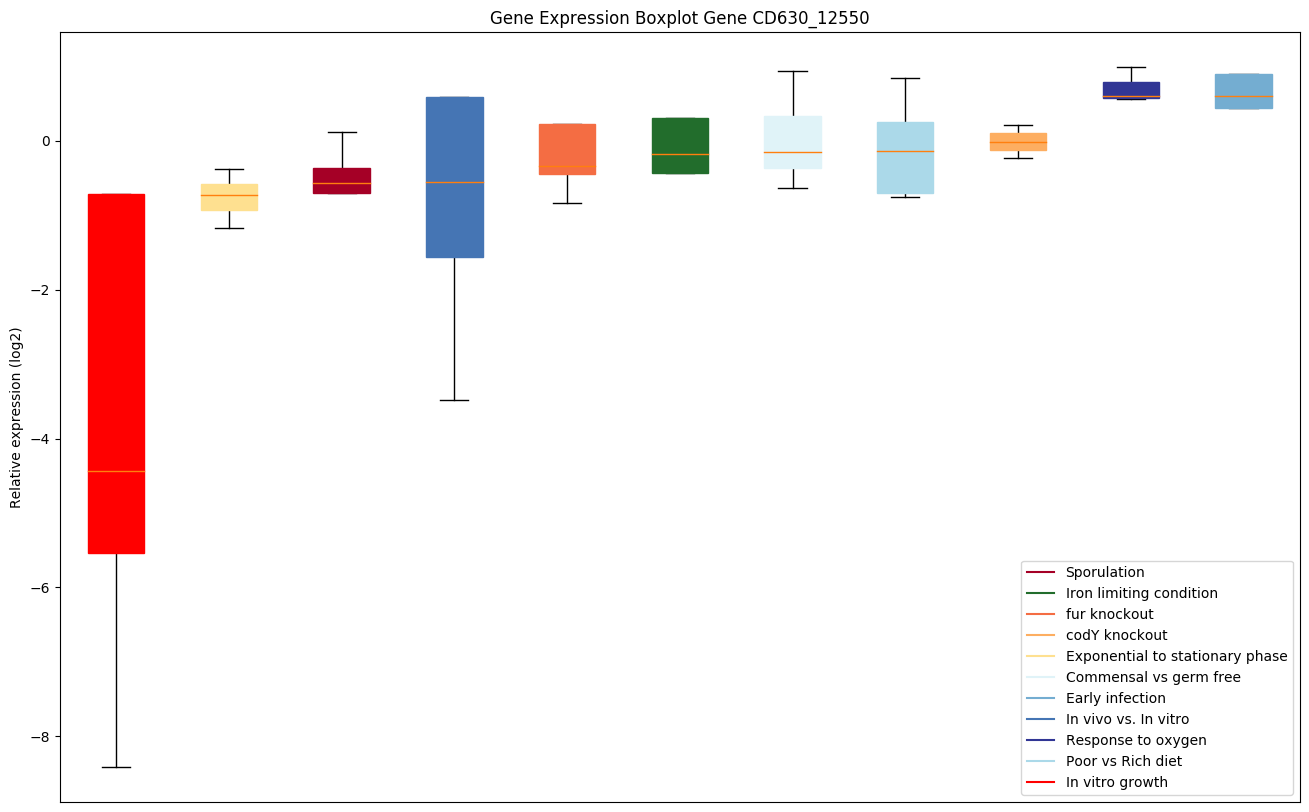
| CD630_12550 | rimM | 16S rRNA processing protein RimM | An accessory protein needed during the final step in the assembly of 30S ribosomal subunit, possibly for assembly of the head region. Probably interacts with S19. Essential for efficient processing of 16S rRNA. May be needed both before and after RbfA during the maturation of 16S rRNA. It has affinity for free ribosomal 30S subunits but not for 70S ribosomes. | Yes |
 |
||
| CD630_12570 | rplS | 50S ribosomal protein L19 | This protein is located at the 30S-50S ribosomal subunit interface and may play a role in the structure and function of the aminoacyl-tRNA binding site. | Yes |
 |
||
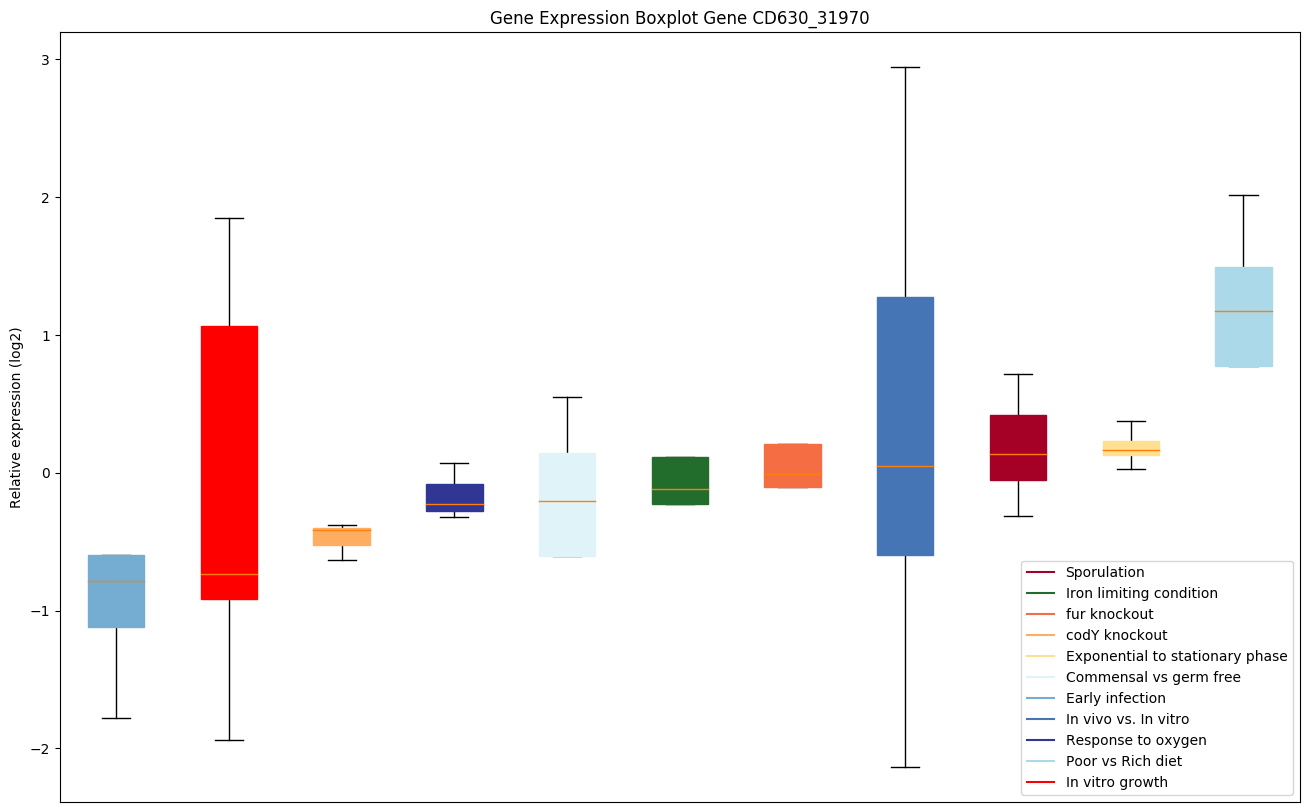
| CD630_31970 | Transcriptional regulator, MerR family |
 |
|||||
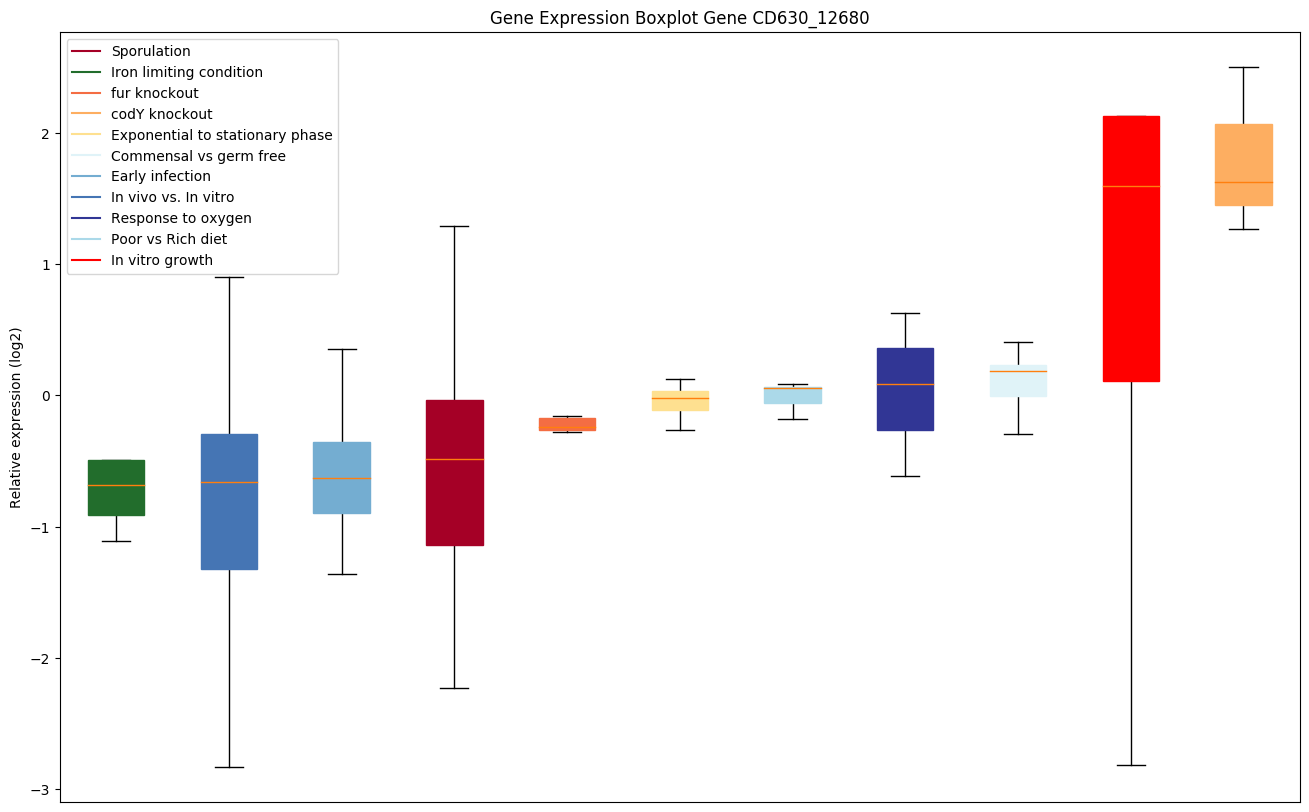
| CD630_12680 | ABC-type transport system, multidrug-familyATP-binding protein |
 |
|||||
| CD630_32610 | pstB | ABC-type transport system, phosphate-specificATP-binding | Part of the ABC transporter complex PstSACB involved in phosphate import. Responsible for energy coupling to the transport system. |
 |
|||
| CD630_31950 | Putative UvrABC system protein A 2 (UvrA protein2) (Excinuclease ABC subunit A 2) |
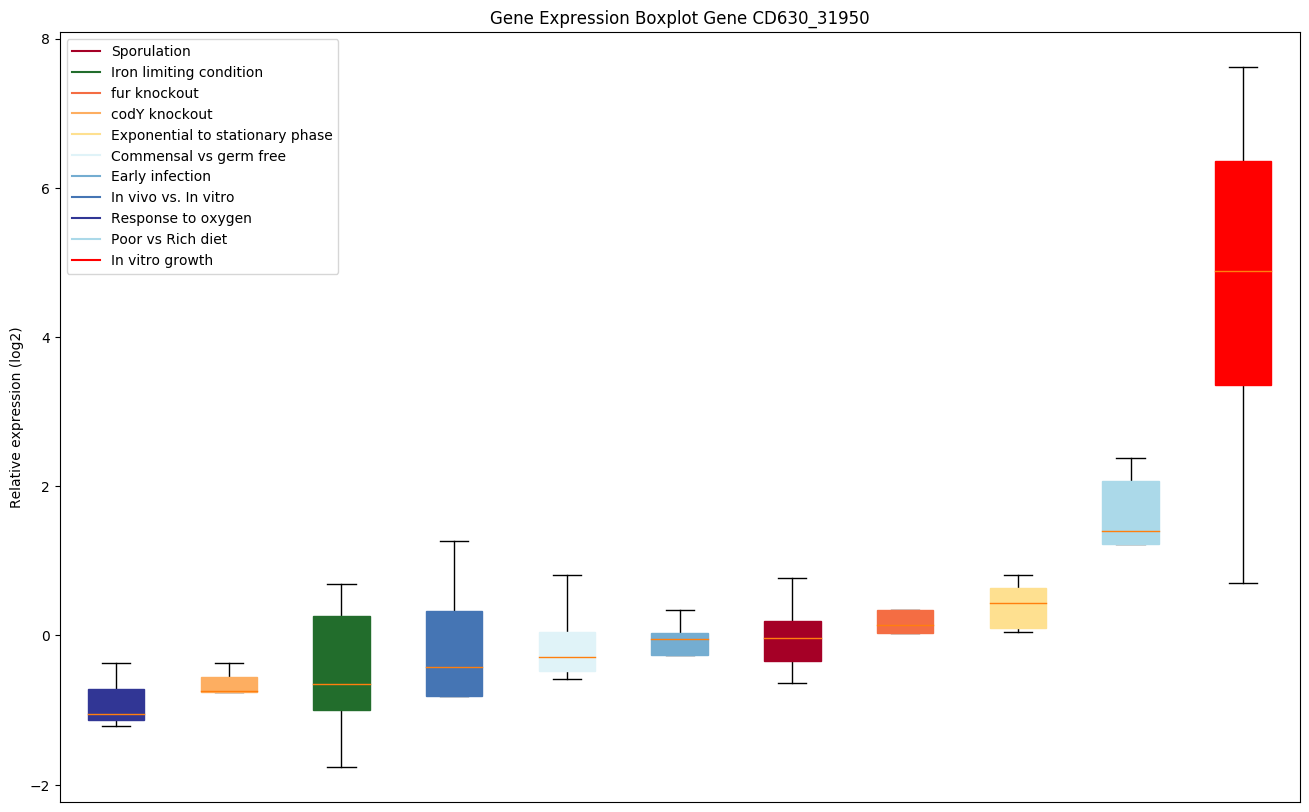 |
|||||
| CD630_32630 | pstC | ABC-type transport system, phosphate-specificpermease | Part of the binding-protein-dependent transport system for phosphate; probably responsible for the translocation of the substrate across the membrane. |
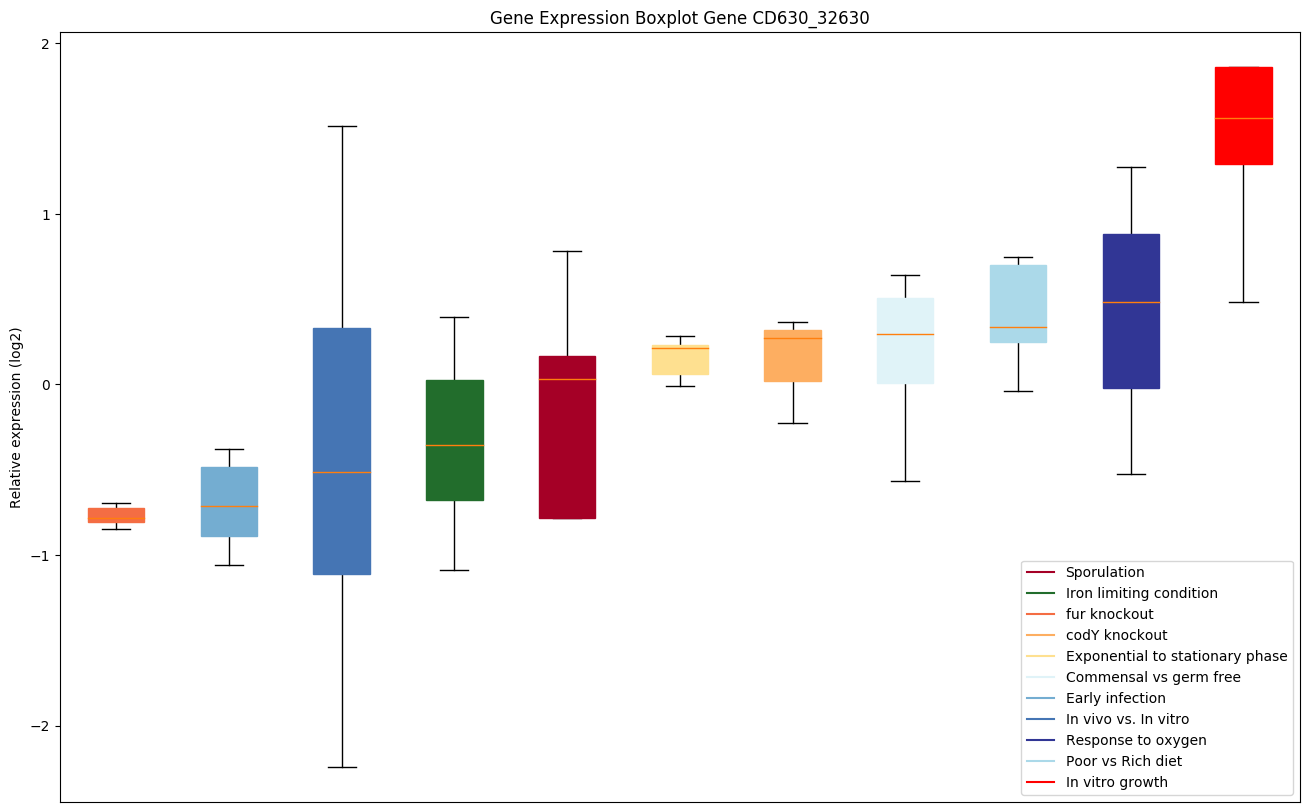 |
|||
| CD630_20180 | Transcriptional regulator, AraC family |
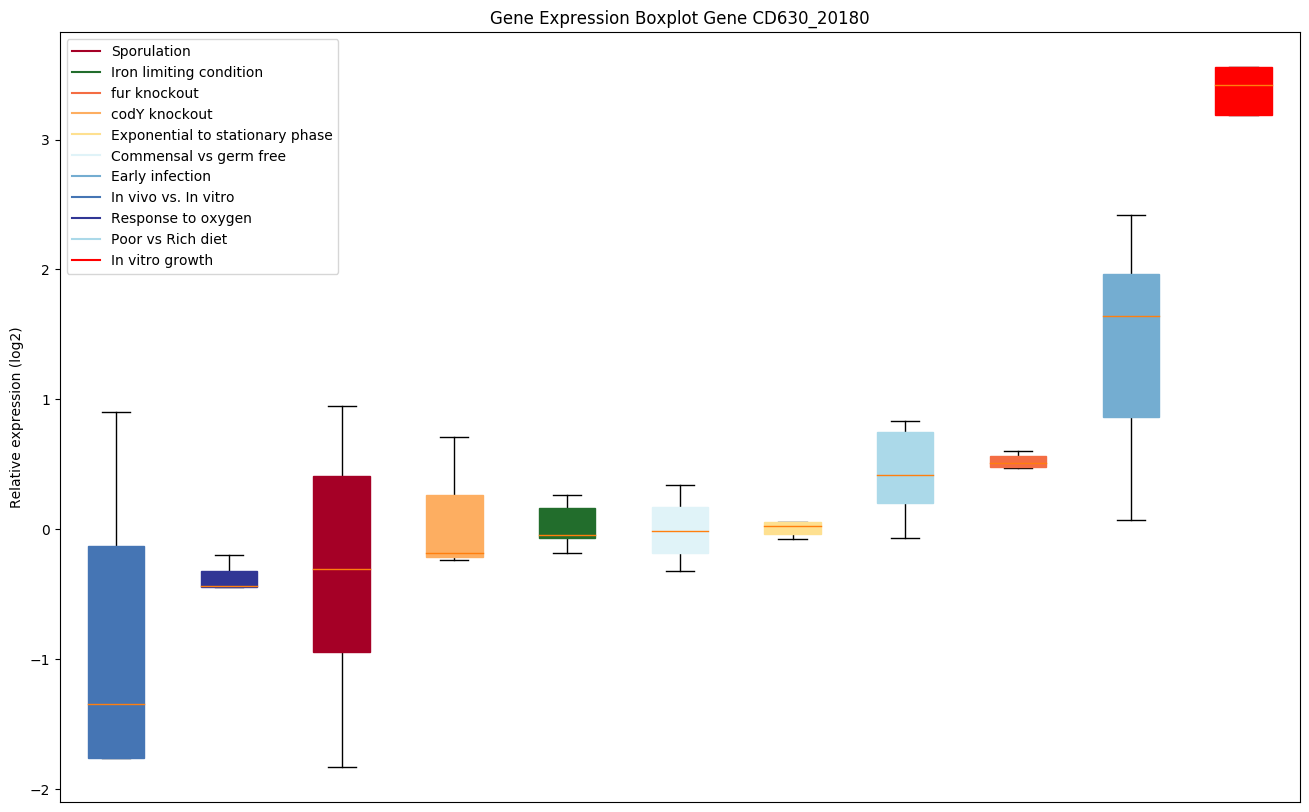 |