Module 28
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.12 | 16 | 62 | NA | NA |
Bicluster expression profile
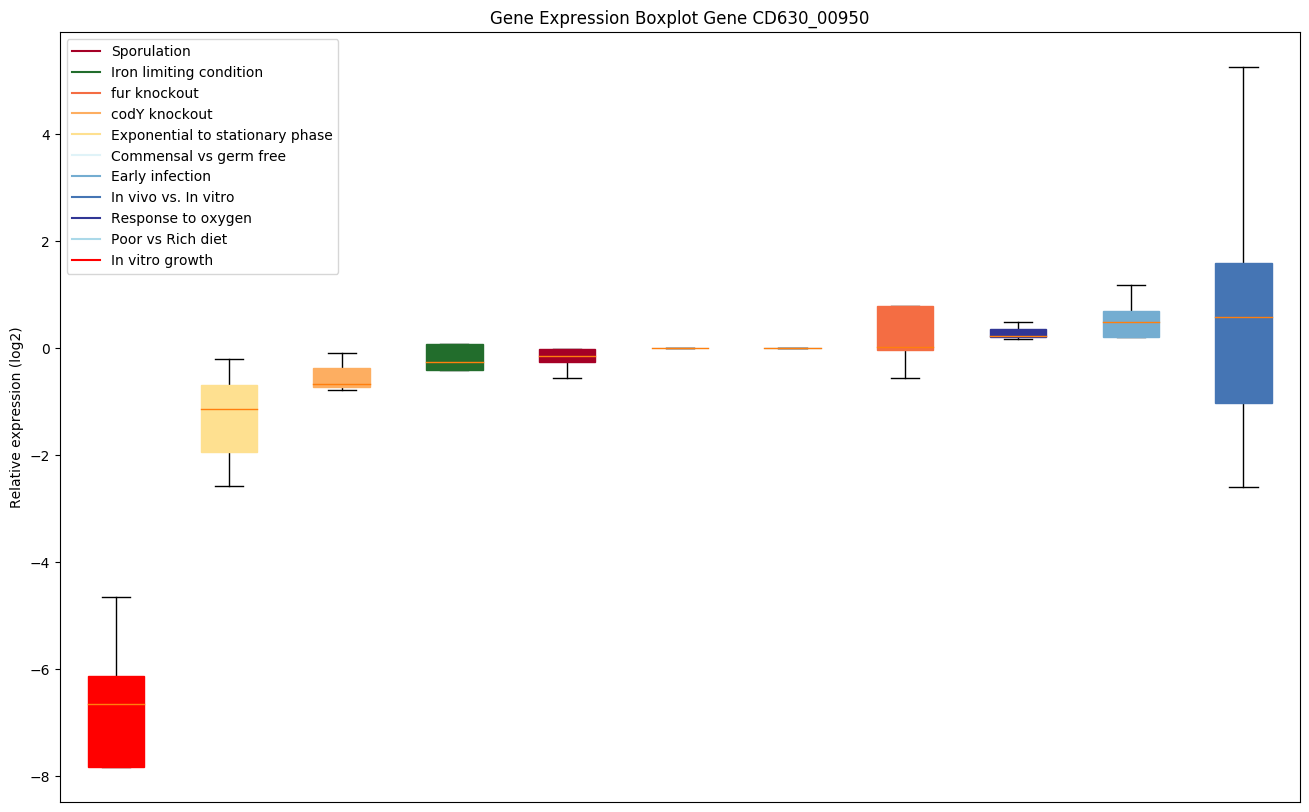
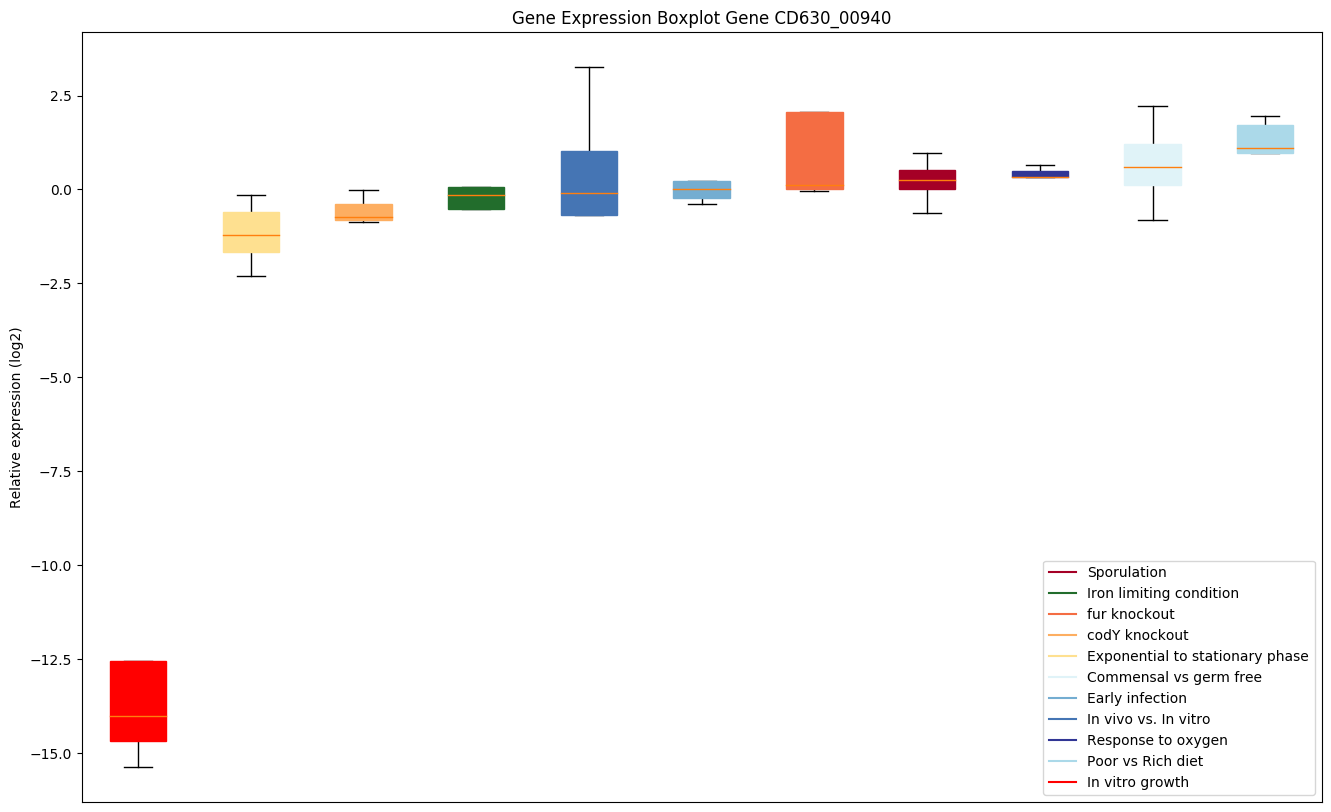
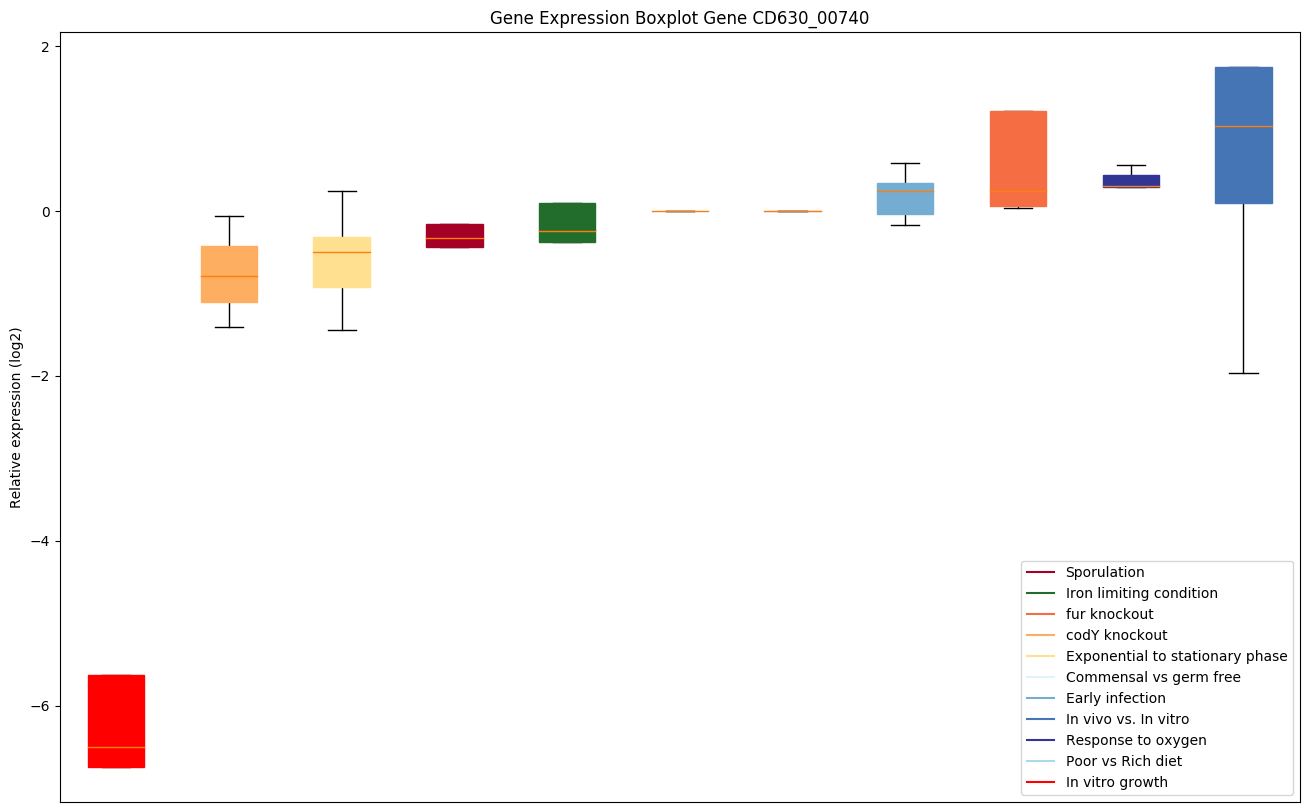
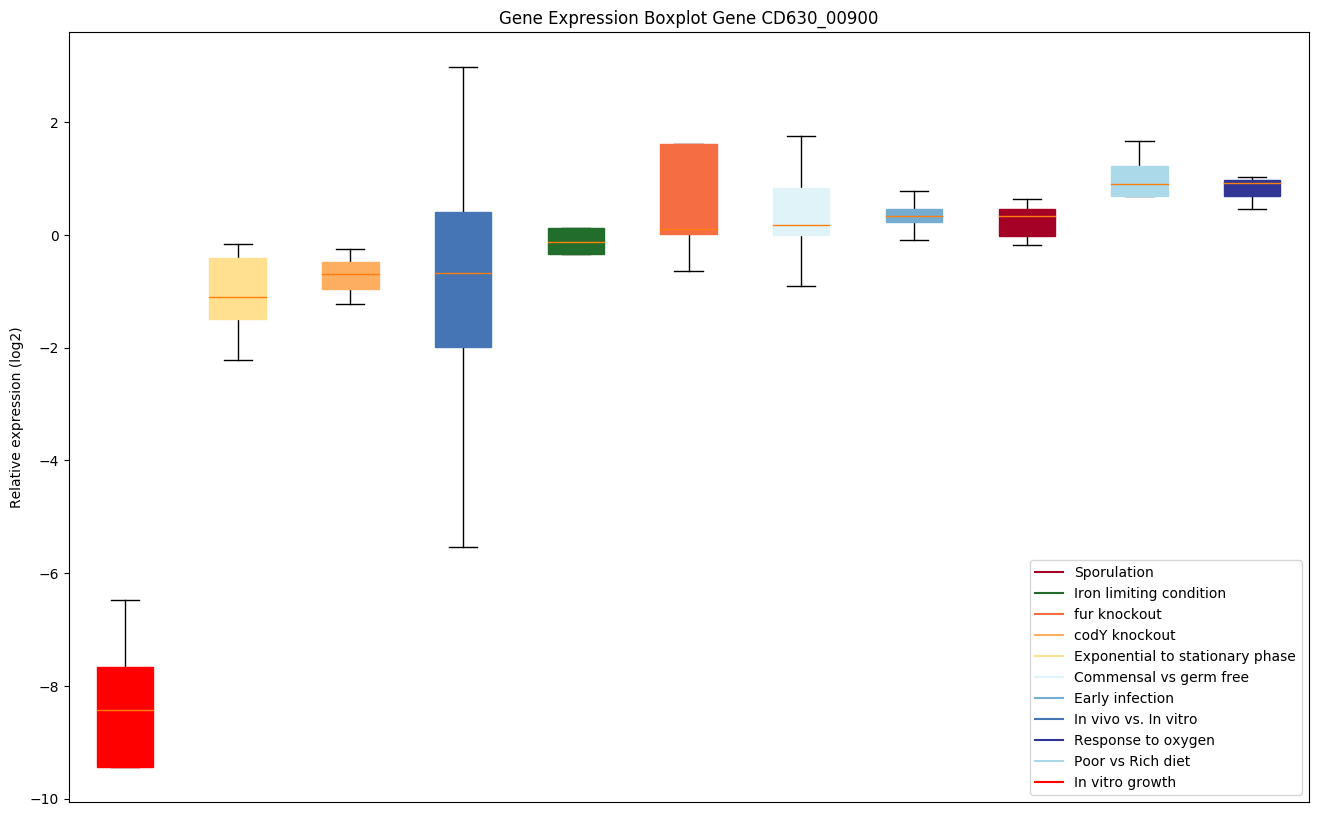
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
| Motif 1 | Motif 1 e-value | Motif 2 | Motif 2 e-value | ||
|---|---|---|---|---|---|
| NA | NA | NA | NA |
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
The module is significantly enriched with genes associated to the indicated functional terms
The enrichment was determined with hypergeometric test using the genome functional annotation compiled by Girinathan et al (2020).
| Module | Pathway | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
| 28 | Ribosome Synthesis and Modification | 11 | 0.000000 | 0.000000 |
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_00780 | rplV | 50S ribosomal protein L22 | This protein binds specifically to 23S rRNA; its binding is stimulated by other ribosomal proteins, e.g. L4, L17, and L20. It is important during the early stages of 50S assembly. It makes multiple contacts with different domains of the 23S rRNA in the assembled 50S subunit and ribosome (By similarity). | Yes |
 |
||
| CD630_00870 | rplR | 50S ribosomal protein L18 | This is one of the proteins that binds and probably mediates the attachment of the 5S RNA into the large ribosomal subunit, where it forms part of the central protuberance. | Yes |
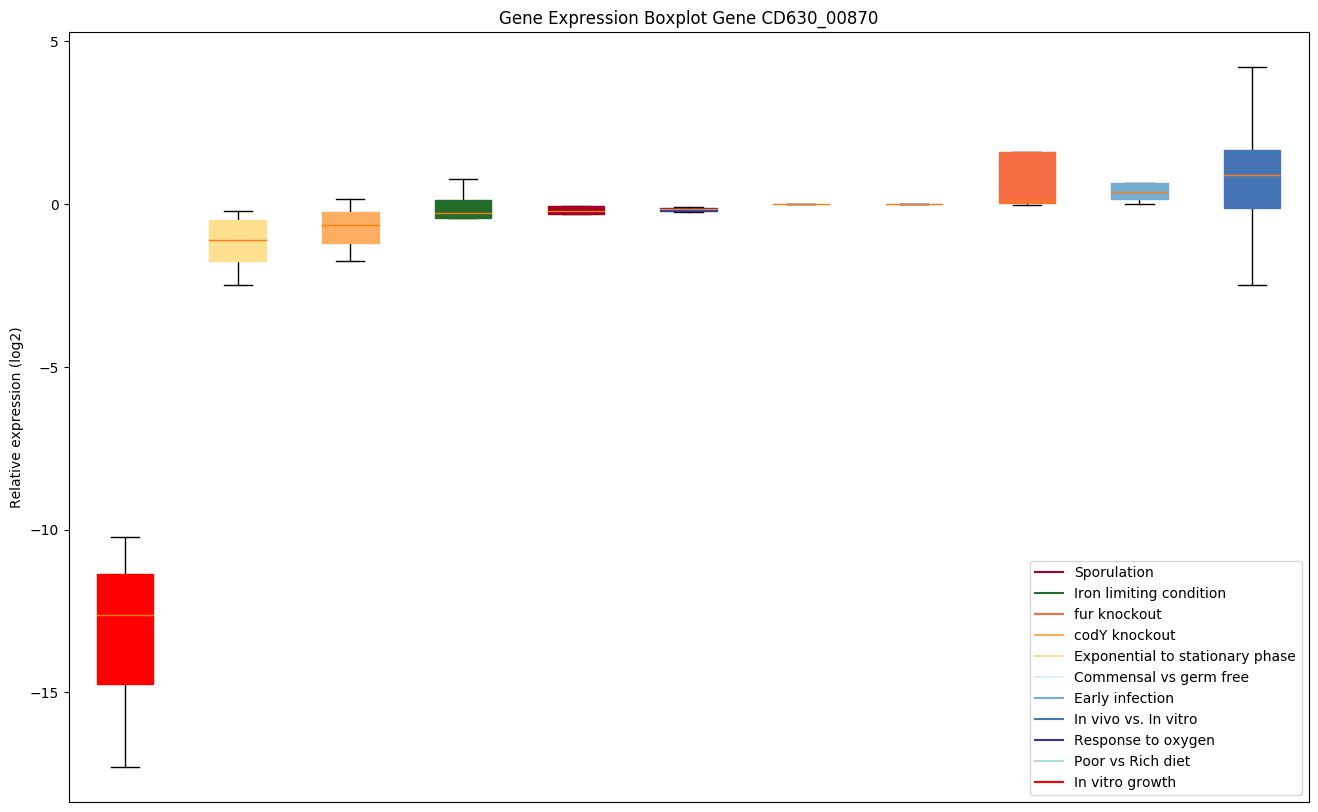 |
||
| CD630_00730 | rplC | 50S ribosomal protein L3 | One of the primary rRNA binding proteins, it binds directly near the 3'-end of the 23S rRNA, where it nucleates assembly of the 50S subunit. | Yes |
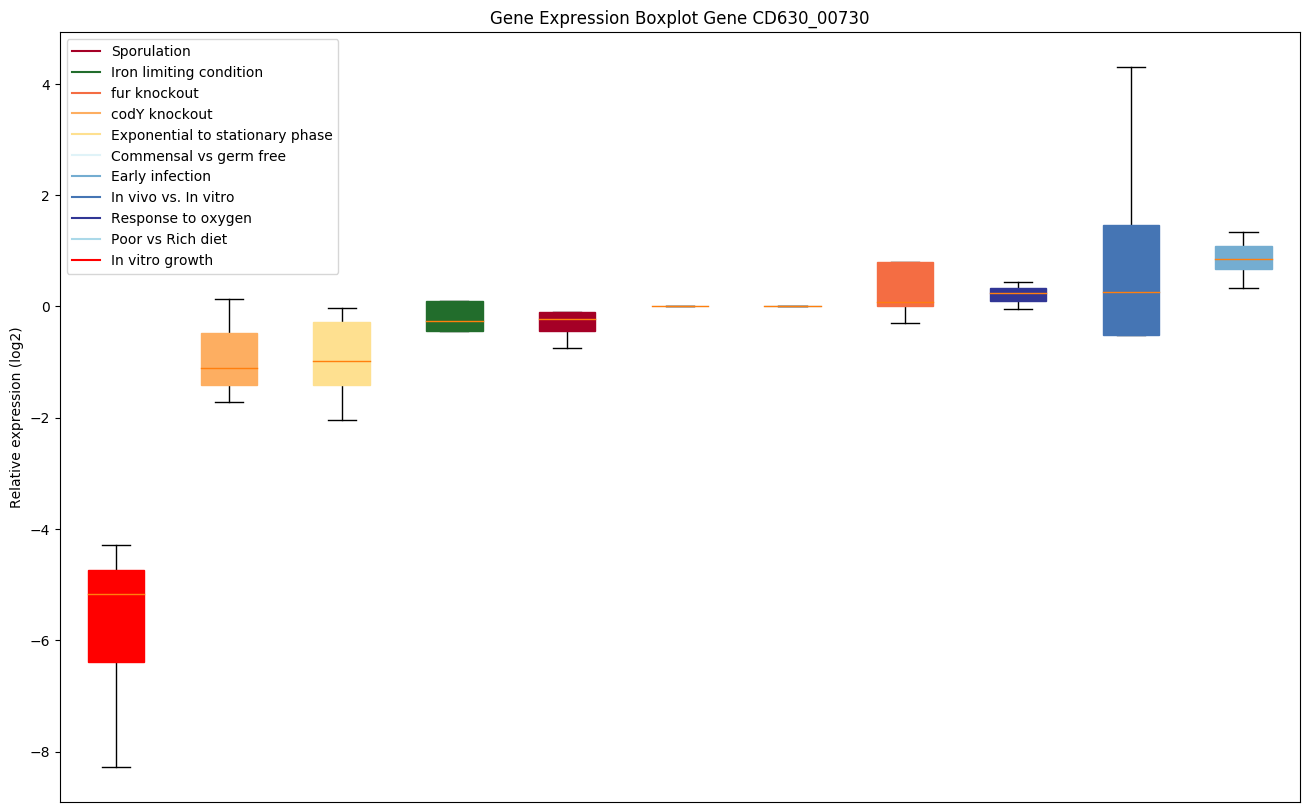 |
||
| CD630_00790 | rpsC | 30S ribosomal protein S3 | Binds the lower part of the 30S subunit head. Binds mRNA in the 70S ribosome, positioning it for translation. | Yes |
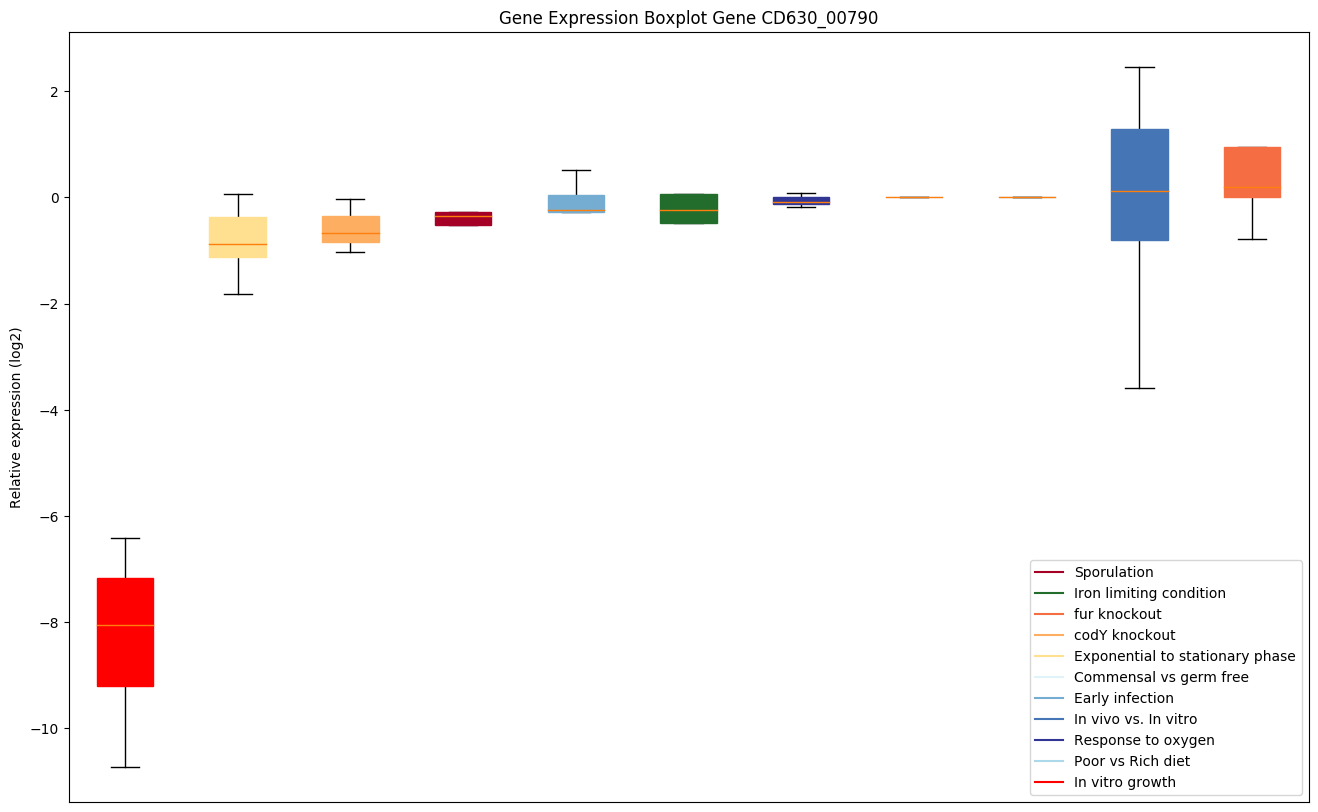 |
||
| CD630_00890 | rplO | 50S ribosomal protein L15 | Binds to the 23S rRNA. | Yes |
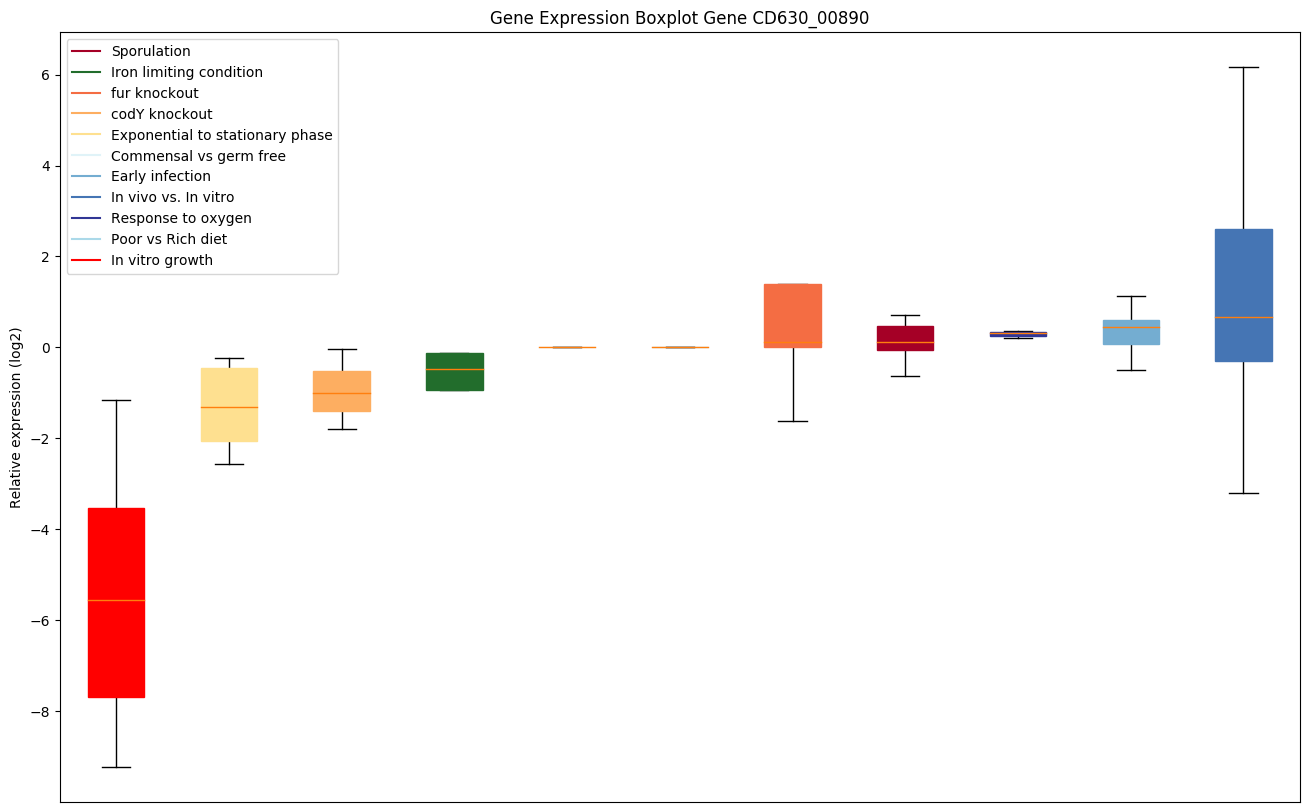 |
||
| CD630_00950 | rpsM | 30S ribosomal protein S13 | Located at the top of the head of the 30S subunit, it contacts several helices of the 16S rRNA. In the 70S ribosome it contacts the 23S rRNA (bridge B1a) and protein L5 of the 50S subunit (bridge B1b), connecting the 2 subunits; these bridges are implicated in subunit movement. Contacts the tRNAs in the A and P-sites. | Yes |
 |
||
| CD630_00940 | infA | Translation initiation factor IF-1 | One of the essential components for the initiation of protein synthesis. Stabilizes the binding of IF-2 and IF-3 on the 30S subunit to which N-formylmethionyl-tRNA(fMet) subsequently binds. Helps modulate mRNA selection, yielding the 30S pre-initiation complex (PIC). Upon addition of the 50S ribosomal subunit IF-1, IF-2 and IF-3 are released leaving the mature 70S translation initiation complex. | Yes |
 |
||
| CD630_00740 | rplD | 50S ribosomal protein L4 | Forms part of the polypeptide exit tunnel. | Yes |
 |
||
| CD630_00900 | prlA | Preprotein translocase SecY subunit | The central subunit of the protein translocation channel SecYEG. Consists of two halves formed by TMs 1-5 and 6-10. These two domains form a lateral gate at the front which open onto the bilayer between TMs 2 and 7, and are clamped together by SecE at the back. The channel is closed by both a pore ring composed of hydrophobic SecY resides and a short helix (helix 2A) on the extracellular side of the membrane which forms a plug. The plug probably moves laterally to allow the channel to open. The ring and the pore may move independently. | Yes |
 |
||
| CD630_01040 | rplM | 50S ribosomal protein L13 | This protein is one of the early assembly proteins of the 50S ribosomal subunit, although it is not seen to bind rRNA by itself. It is important during the early stages of 50S assembly. |
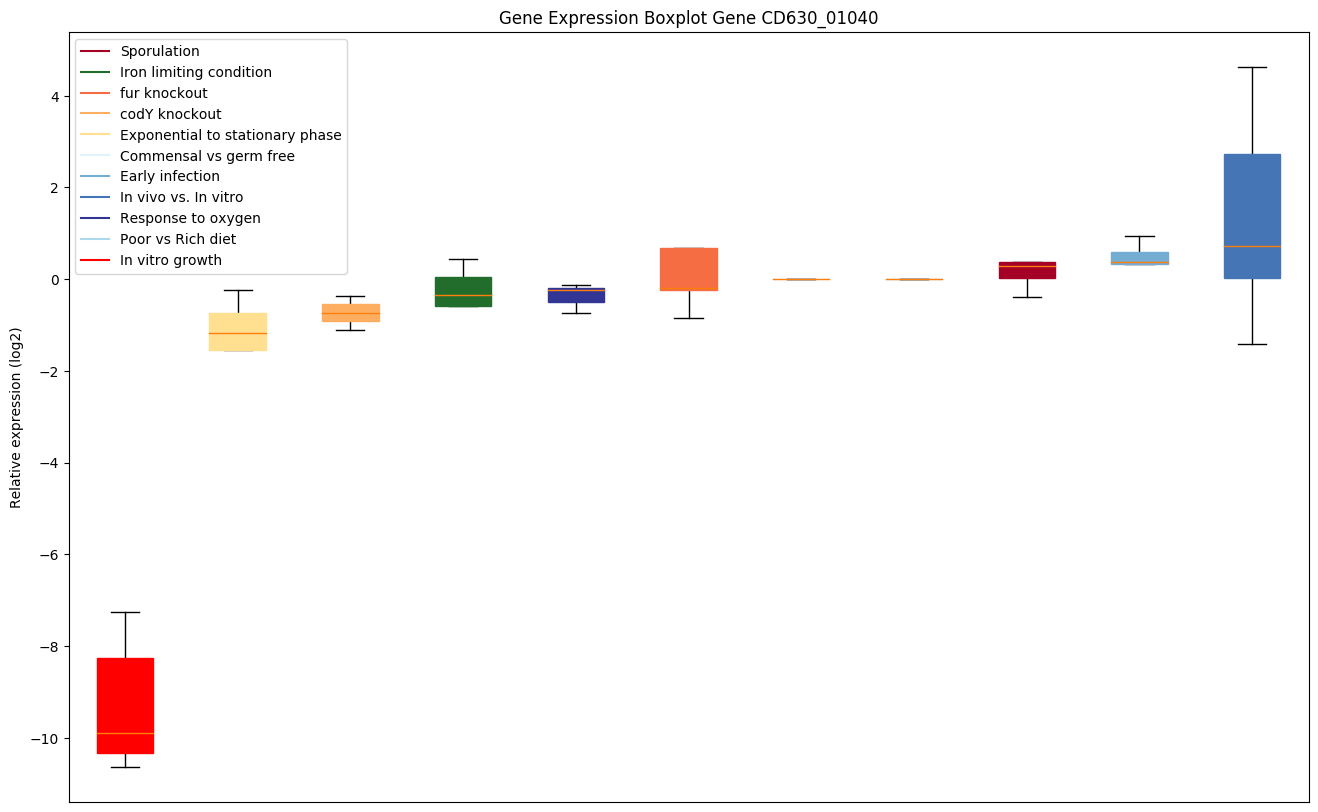 |
|||
| CD630_00770 | rpsS | 30S ribosomal protein S19 | Protein S19 forms a complex with S13 that binds strongly to the 16S ribosomal RNA. | Yes |
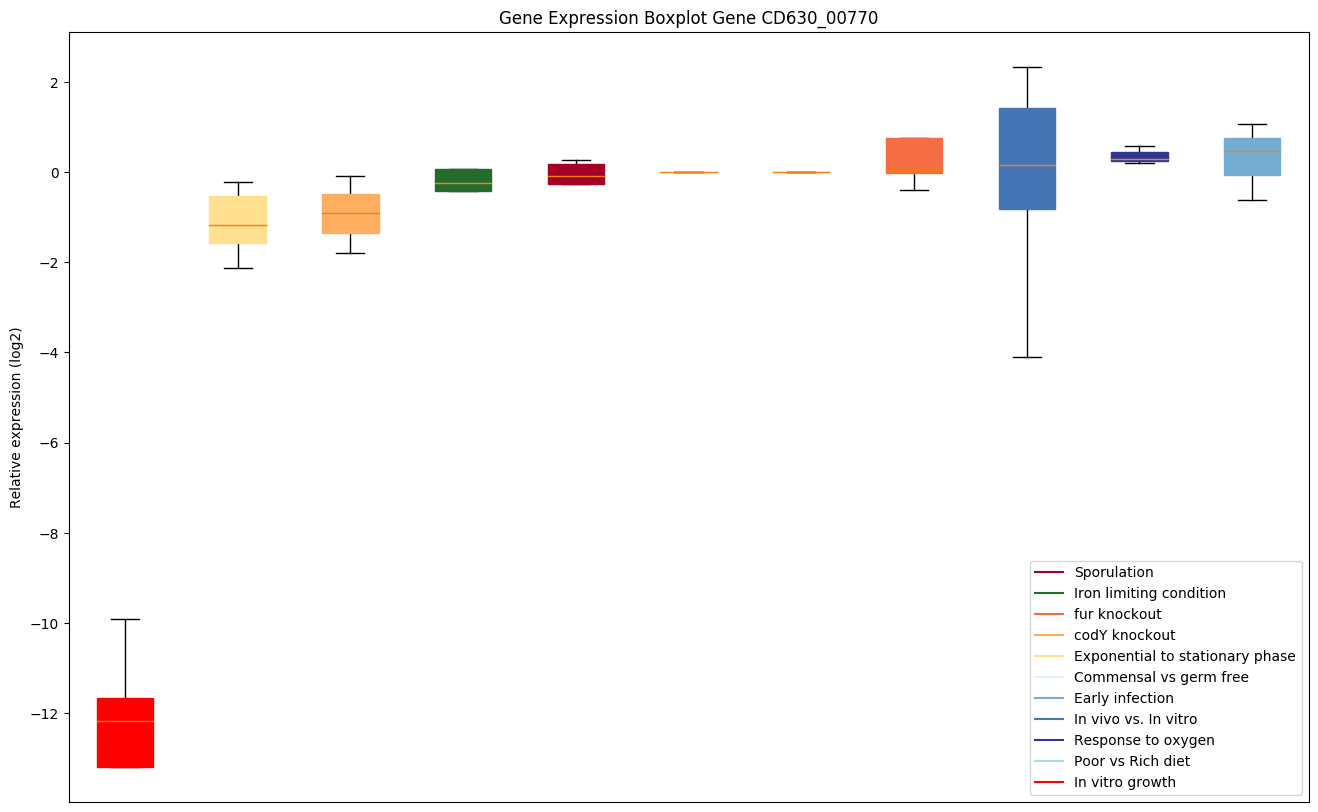 |
||
| CD630_00700 | fusA | Elongation factor G (EF-G) | Catalyzes the GTP-dependent ribosomal translocation step during translation elongation. During this step, the ribosome changes from the pre-translocational (PRE) to the post-translocational (POST) state as the newly formed A-site-bound peptidyl-tRNA and P-site-bound deacylated tRNA move to the P and E sites, respectively. Catalyzes the coordinated movement of the two tRNA molecules, the mRNA and conformational changes in the ribosome. | Yes |
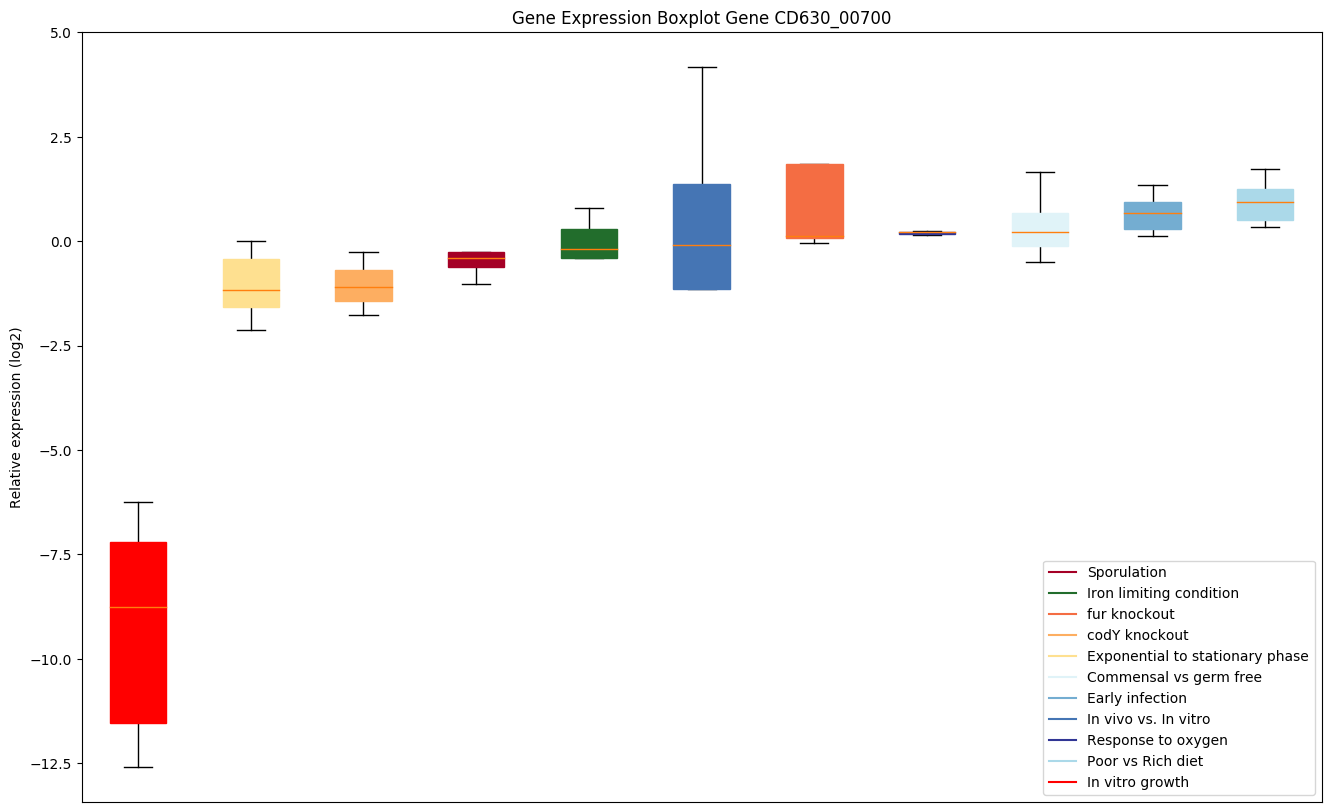 |
||
| CD630_00920 | map1 | Methionine aminopeptidase Map1 (MAP) (PeptidaseM) | Removes the N-terminal methionine from nascent proteins. The N-terminal methionine is often cleaved when the second residue in the primary sequence is small and uncharged (Met-Ala-, Cys, Gly, Pro, Ser, Thr, or Val). Requires deformylation of the N(alpha)-formylated initiator methionine before it can be hydrolyzed. | Yes |
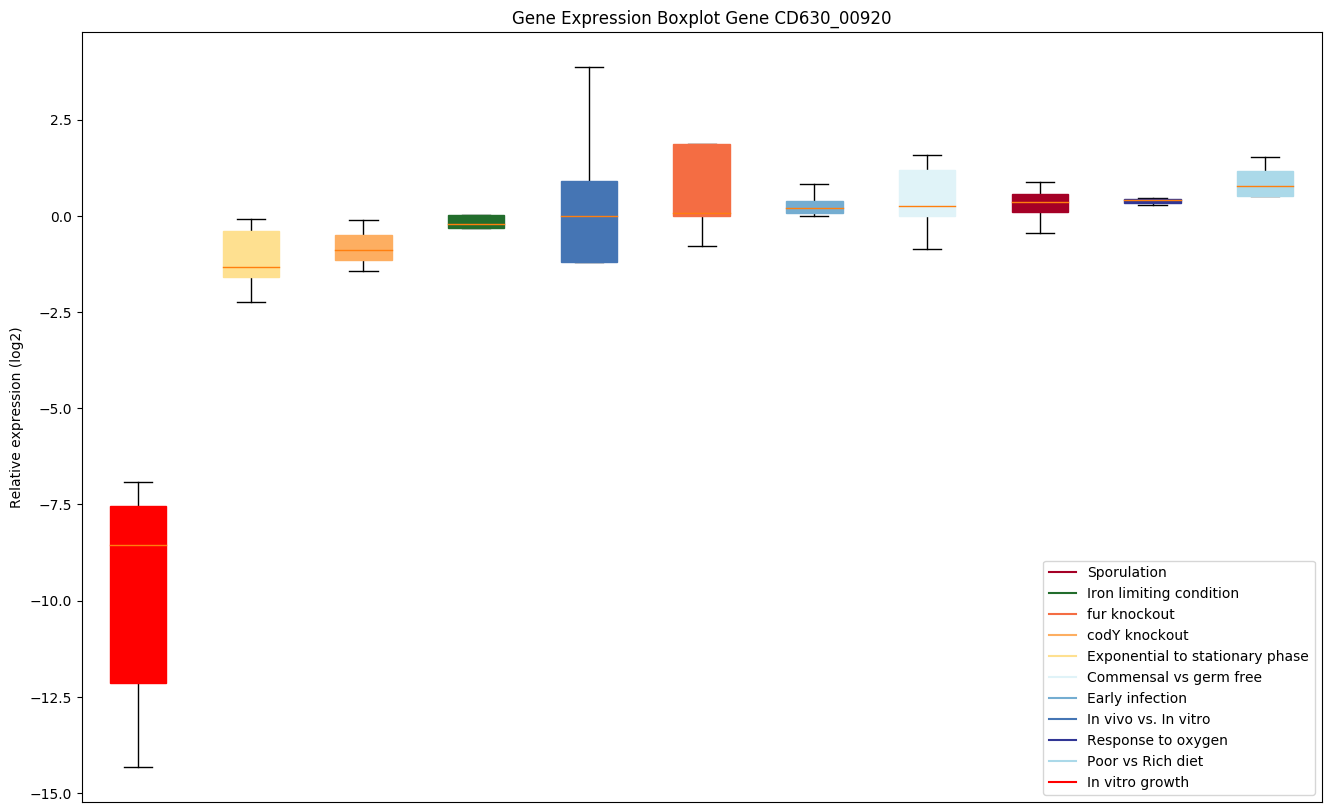 |
||
| CD630_00910 | adk | Adenylate kinase | Catalyzes the reversible transfer of the terminal phosphate group between ATP and AMP. Plays an important role in cellular energy homeostasis and in adenine nucleotide metabolism. | Yes |
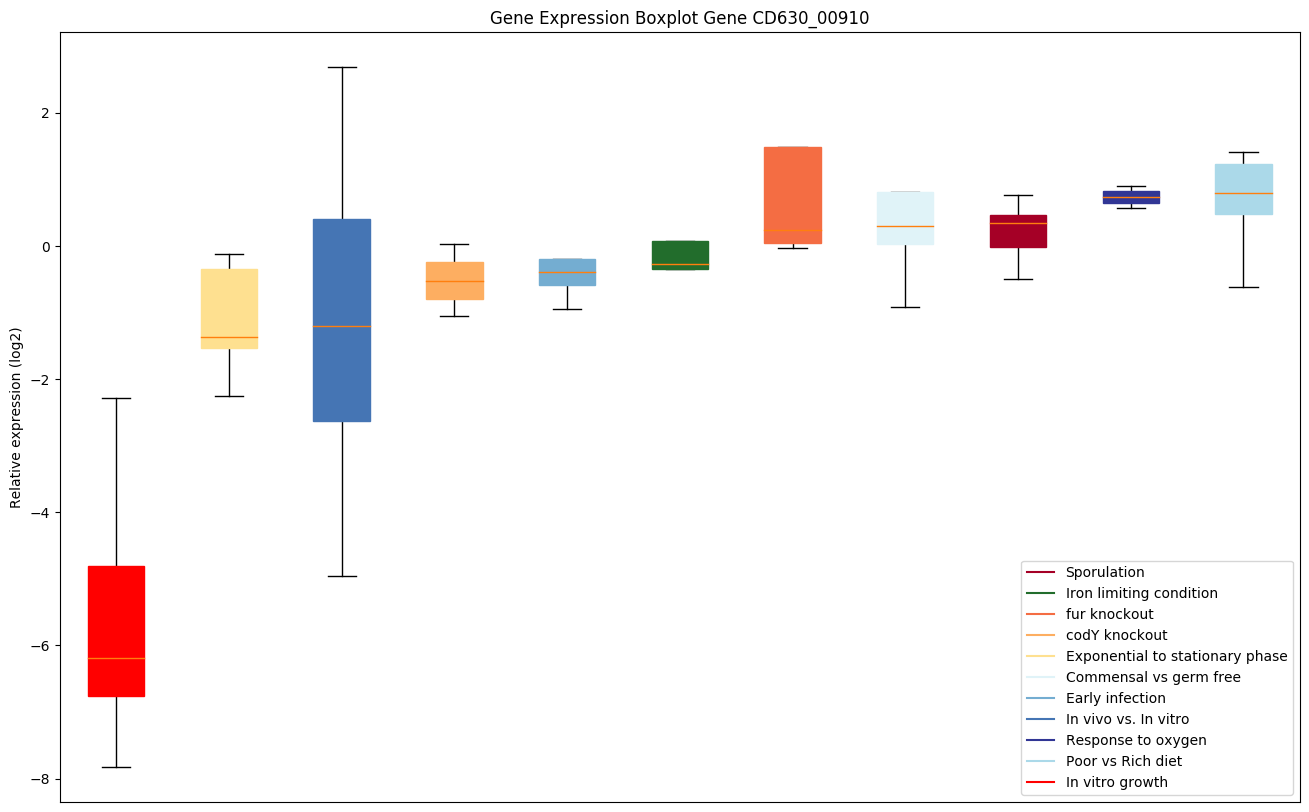 |
||
| CD630_00820 | rplN | 50S ribosomal protein L14 | Binds to 23S rRNA. Forms part of two intersubunit bridges in the 70S ribosome. |
 |
|||
| CD630_00930 | Ribosomal protein L14E/L6E/L27E-like | Yes |
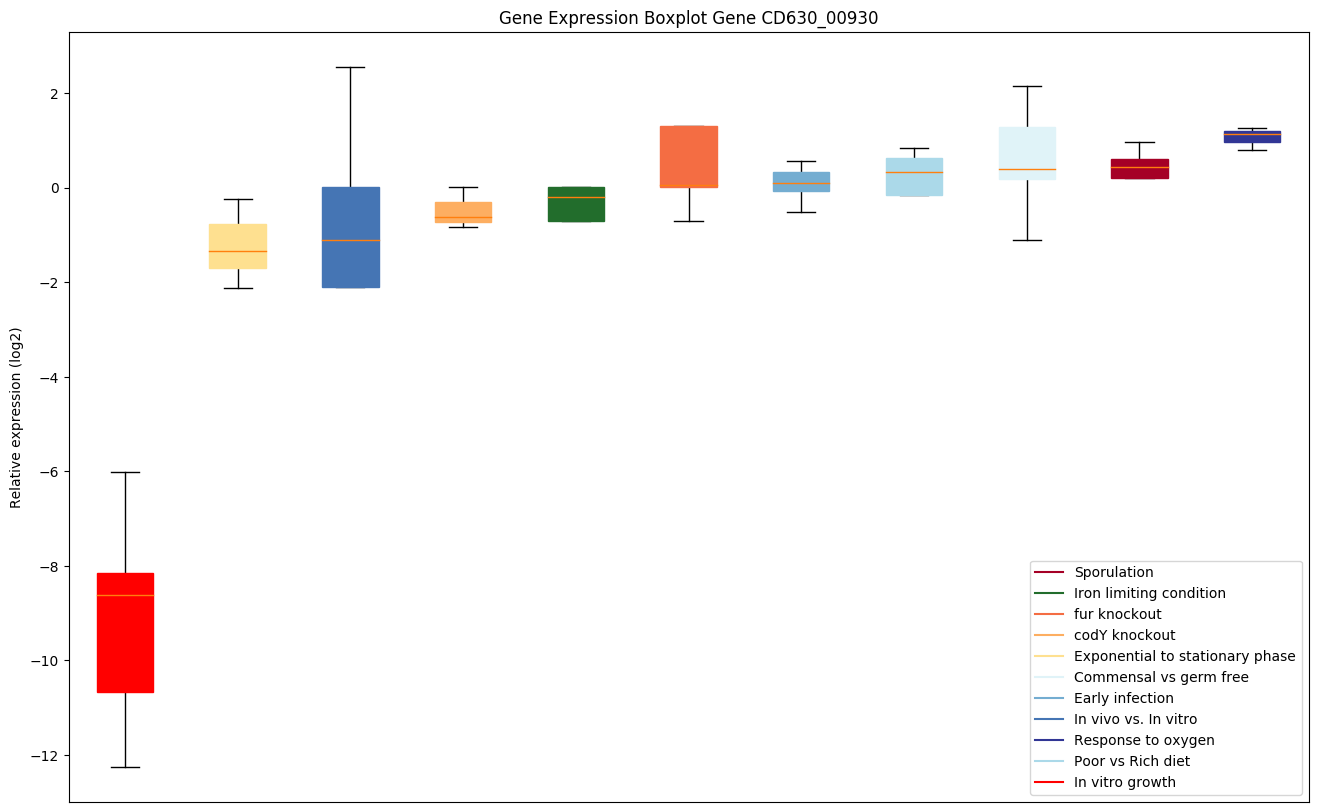 |
