Module 318
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.58 | 21 | 62 | NA | In vivo vs. In vitro |
Bicluster expression profile
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
| TF | Module | Confidence Score | Beta |
|---|---|---|---|
|
cggR Transcriptional regulator, SorC family |
318 | 1.00 | 0.9383 |
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
| TF | Module | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
|
prdR Transcriptional regulator, sigma-54-dependent |
318 | 5 | 0.00 | 0.03 |
The module is significantly enriched with genes associated to the indicated functional terms
The enrichment was determined with hypergeometric test using the genome functional annotation compiled by Girinathan et al (2020).
| Module | Pathway | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
| 318 | CRISPR system | 8 | 0.000000 | 0.000000 |
| 318 | Glycolysis | 5 | 0.000000 | 0.000058 |
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
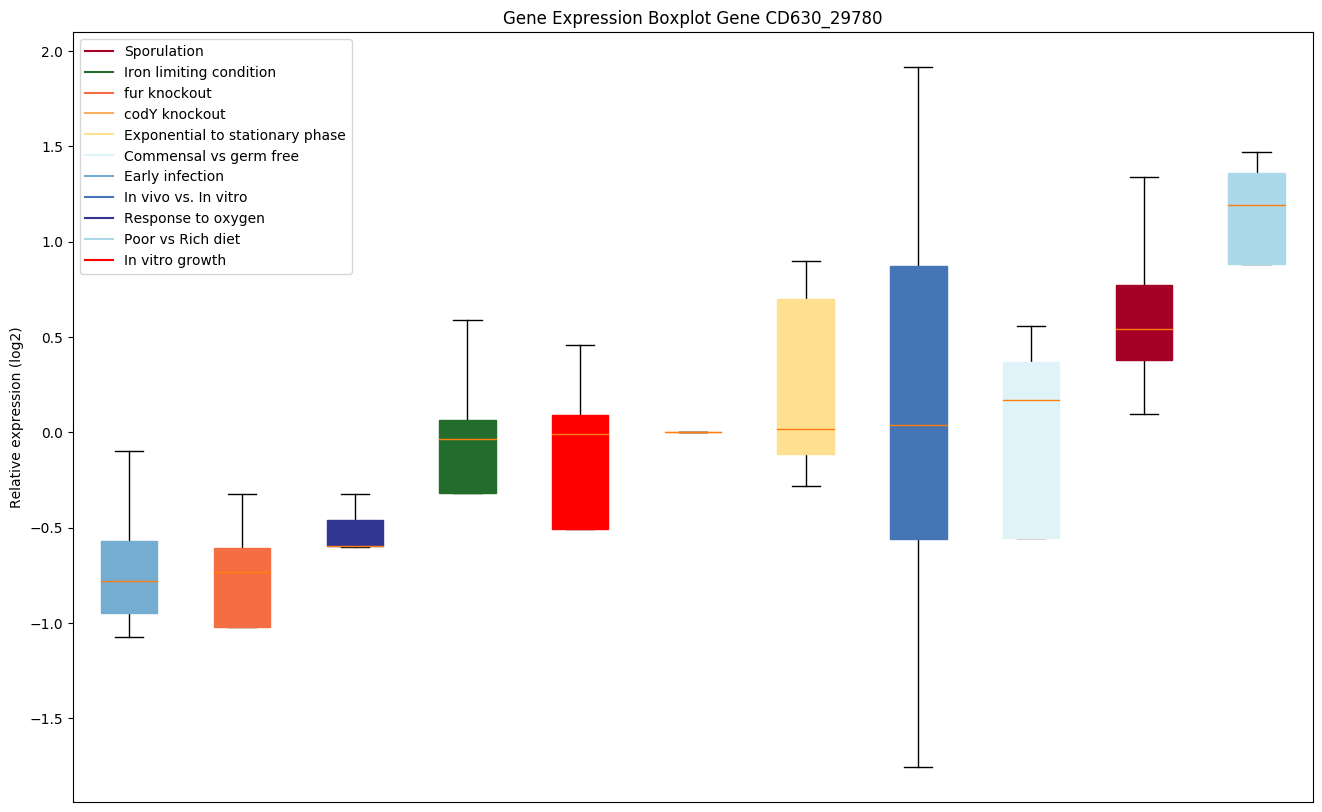
| CD630_29780 | Putative CRISPR-associated Cas3 family helicase |
 |
|||||
| CD630_07150 | ptb | Phosphate butyryltransferase(Phosphotransbutyrylase) |
 |
||||
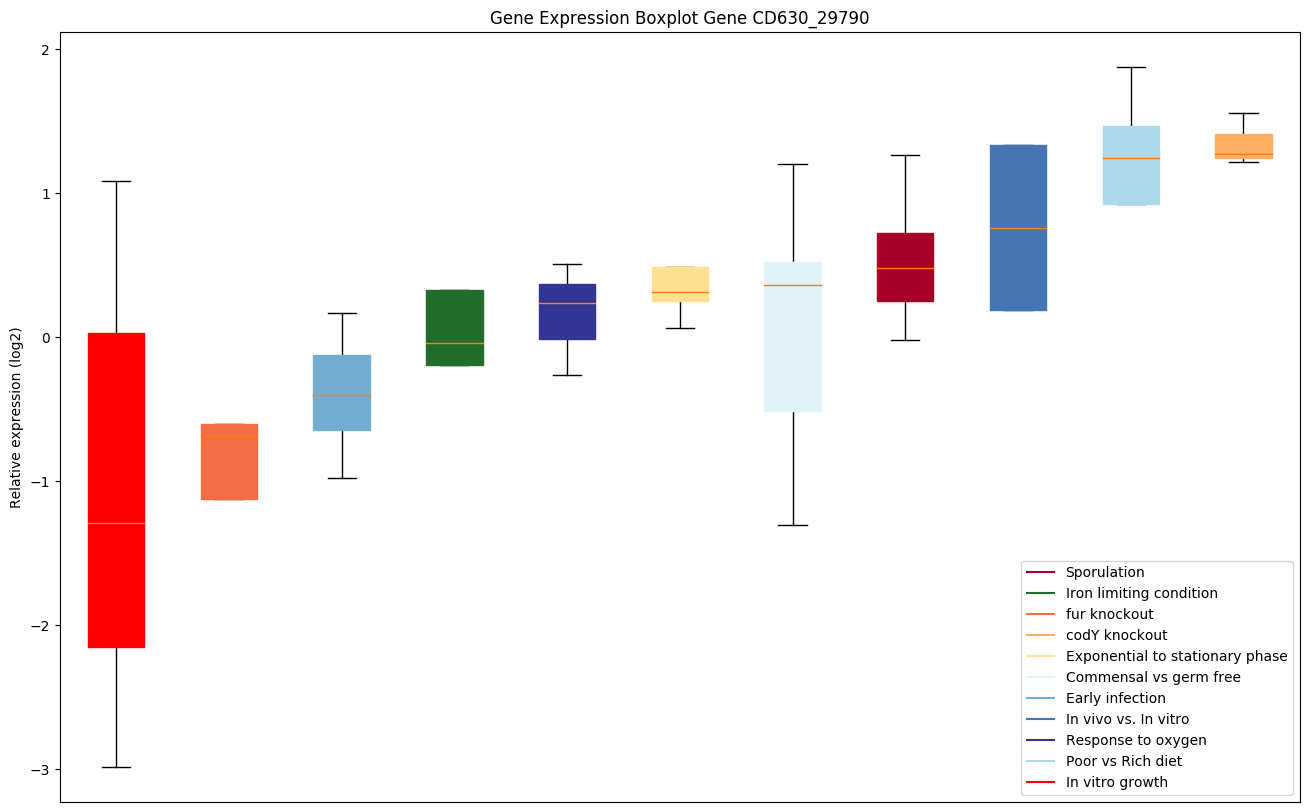
| CD630_29790 | Putative CRISPR-associated Cas5 family protein |
 |
|||||
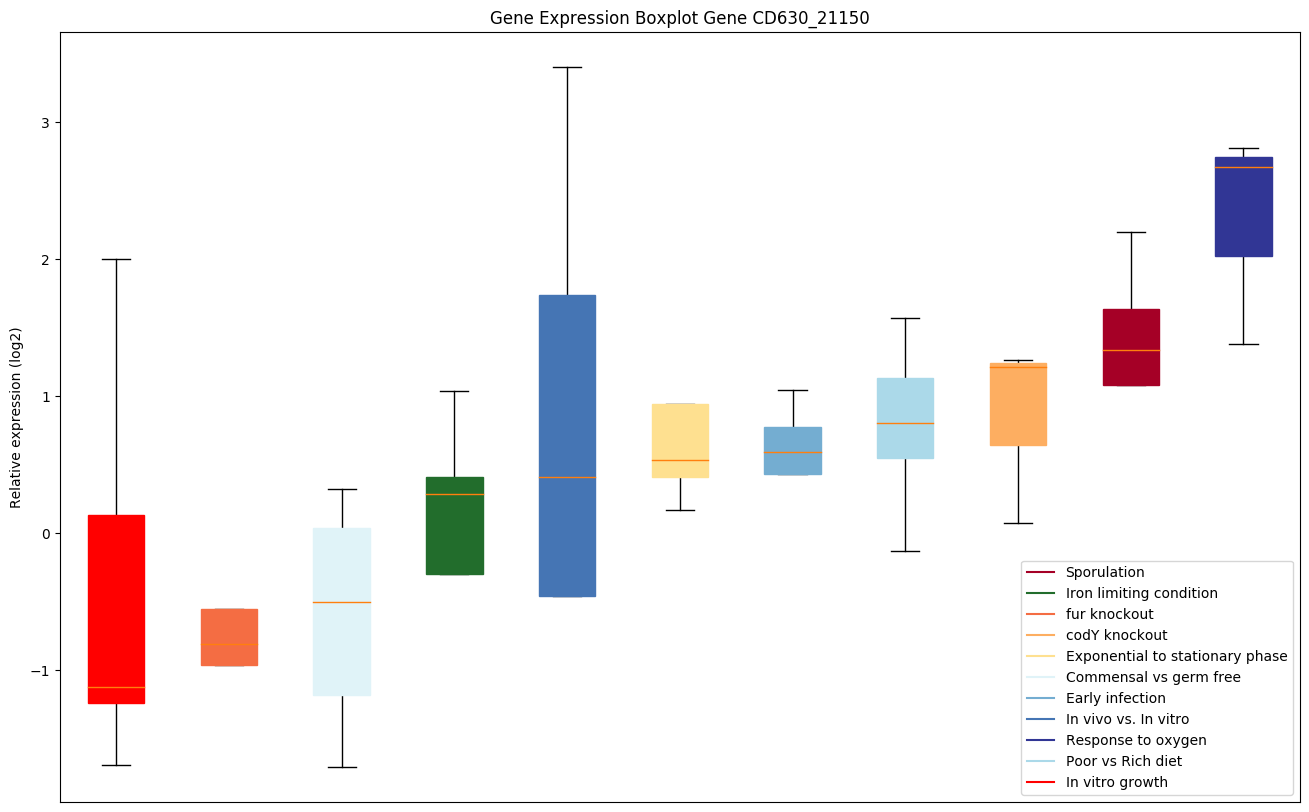
| CD630_21150 | Putative copper-transporting P-type ATPase |
 |
|||||
| CD630_29820 | Putative CRISPR-associated Cas6 family protein |
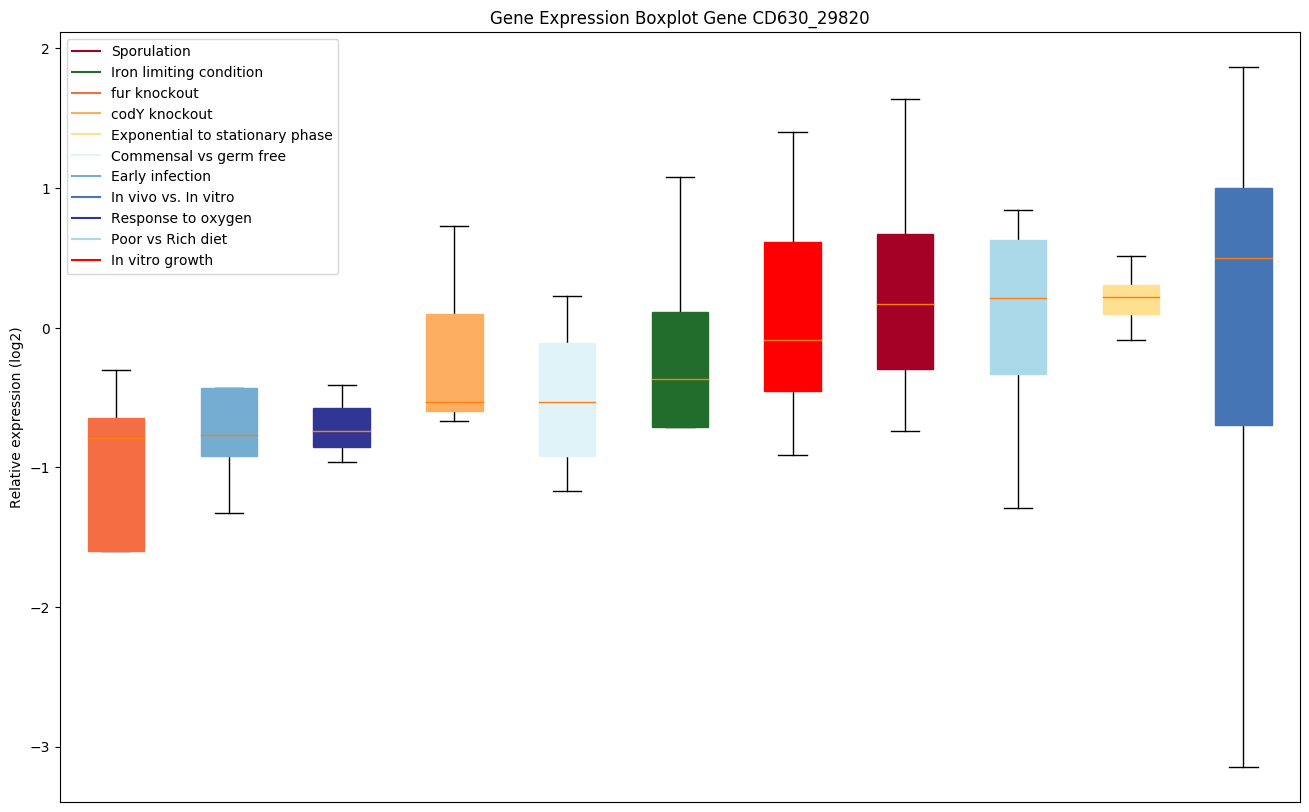 |
|||||
| CD630_27500 | agrB | Accessory gene regulator |
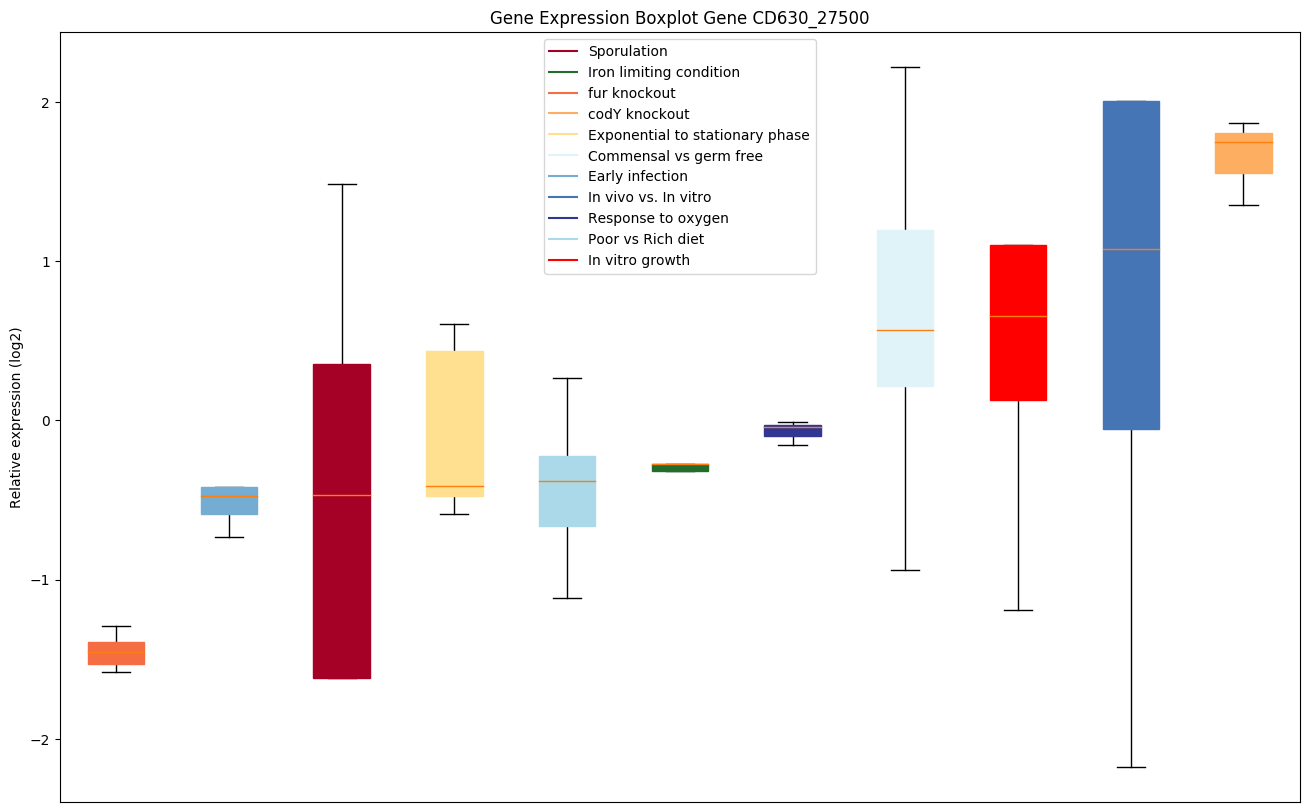 |
||||
| CD630_29770 | Putative CRISPR-associated Cas4 family protein | CRISPR (clustered regularly interspaced short palindromic repeat) is an adaptive immune system that provides protection against mobile genetic elements (viruses, transposable elements and conjugative plasmids). CRISPR clusters contain sequences complementary to antecedent mobile elements and target invading nucleic acids. CRISPR clusters are transcribed and processed into CRISPR RNA (crRNA). |
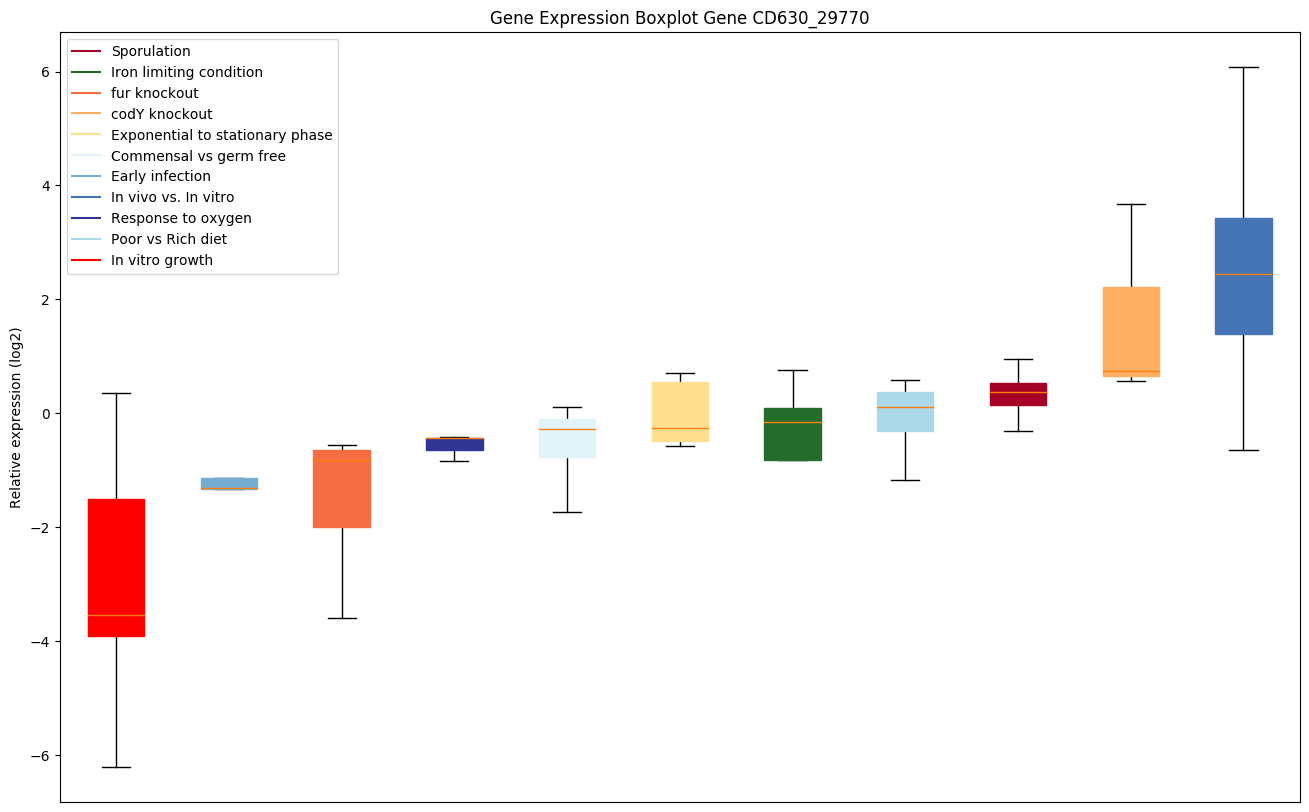 |
||||
| CD630_31720 | tpi | Triosephosphate isomerase (TIM) (Triose-phosphateisomerase) | Involved in the gluconeogenesis. Catalyzes stereospecifically the conversion of dihydroxyacetone phosphate (DHAP) to D-glyceraldehyde-3-phosphate (G3P). | Yes |
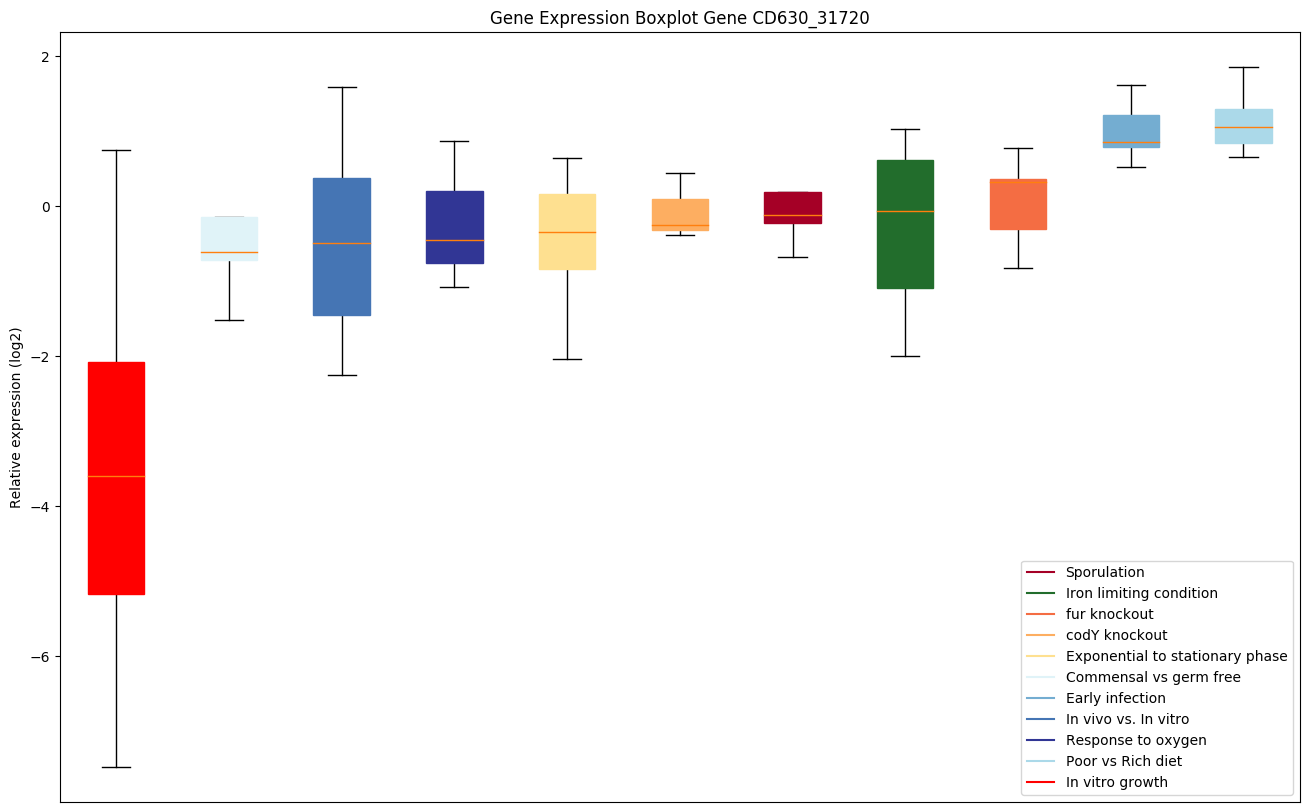 |
||
| CD630_31710 | gpmI | 2,3-bisphosphoglycerate-independentphosphoglycerate mutase (Phosphoglyceromutase)(BPG-independent PGAM) (iPGM) | Catalyzes the interconversion of 2-phosphoglycerate and 3-phosphoglycerate. | Yes |
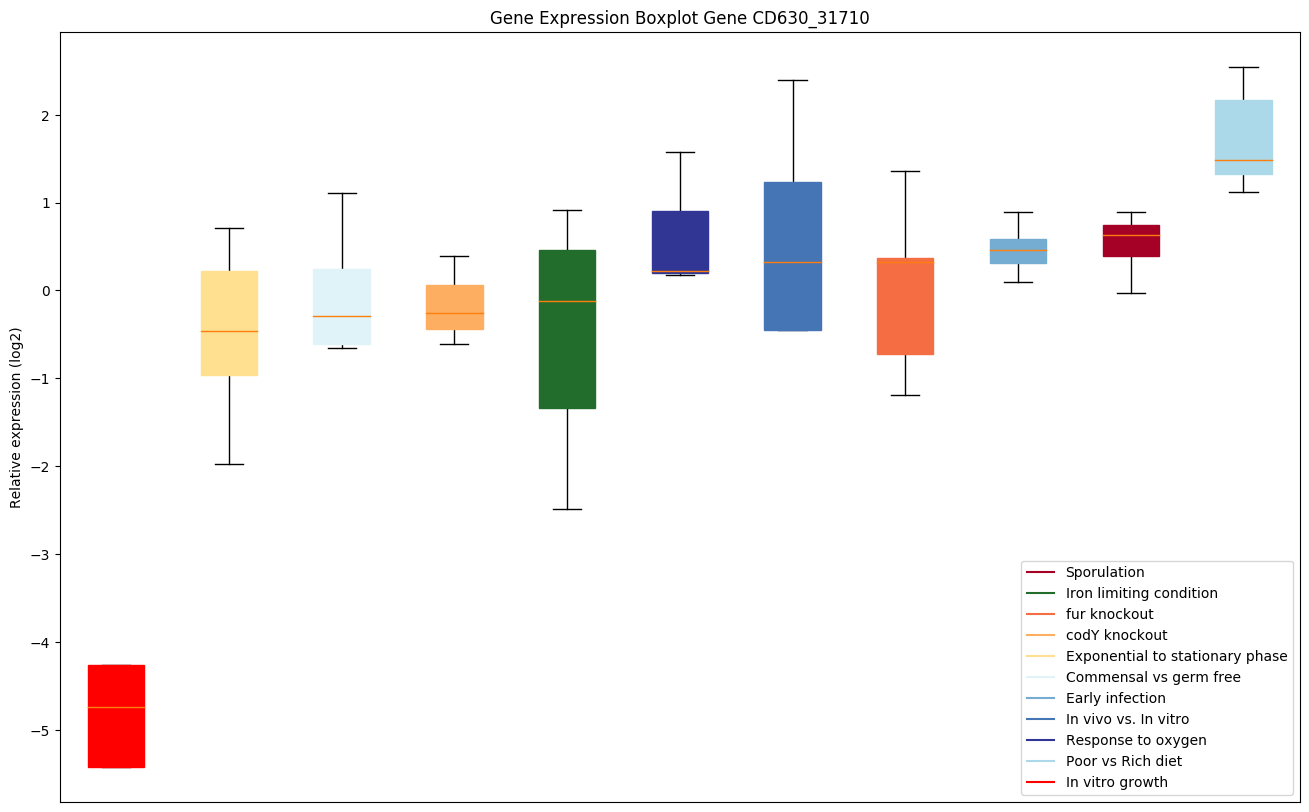 |
||
| CD630_29760 | Putative CRISPR-associated Cas1 family protein | CRISPR (clustered regularly interspaced short palindromic repeat), is an adaptive immune system that provides protection against mobile genetic elements (viruses, transposable elements and conjugative plasmids). CRISPR clusters contain spacers, sequences complementary to antecedent mobile elements, and target invading nucleic acids. CRISPR clusters are transcribed and processed into CRISPR RNA (crRNA). Acts as a dsDNA endonuclease. Involved in the integration of spacer DNA into the CRISPR cassette. |
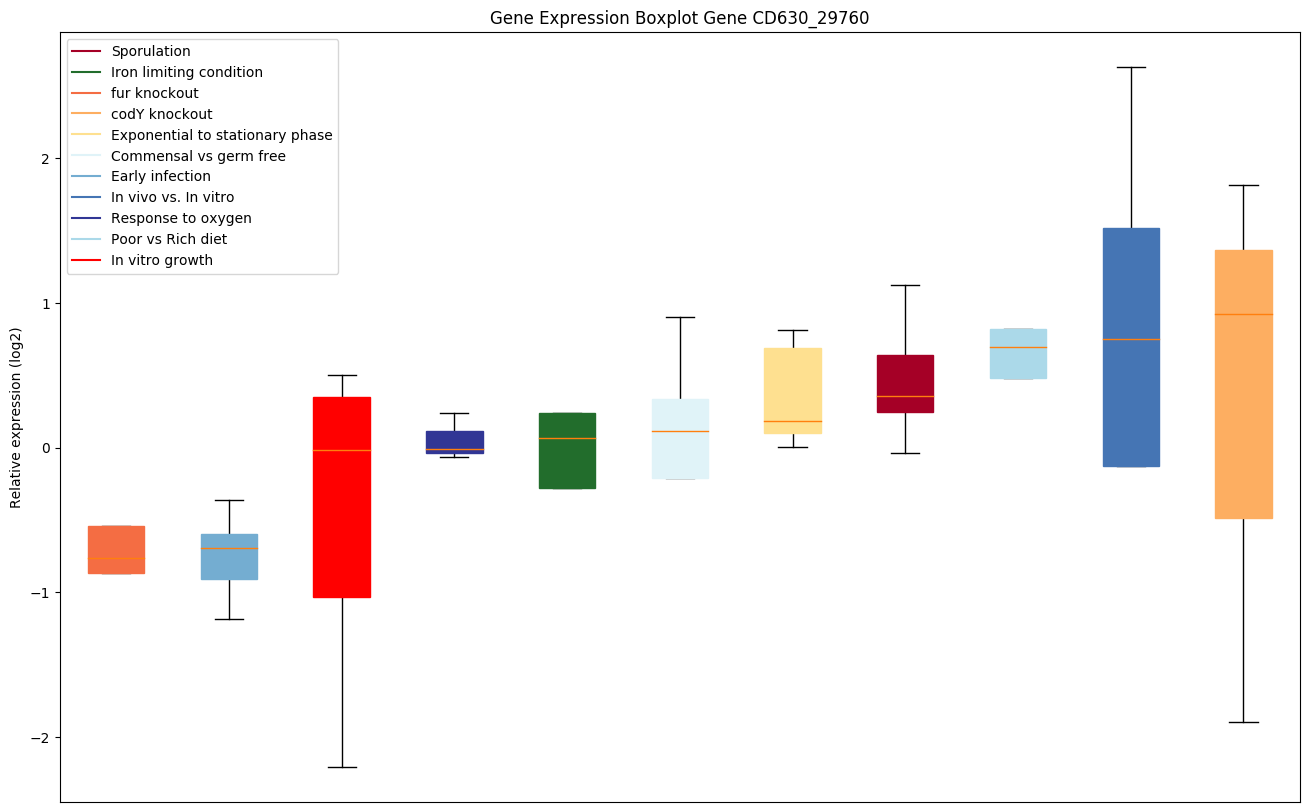 |
||||
| CD630_27491 | agrD | Autoinducer prepeptide |
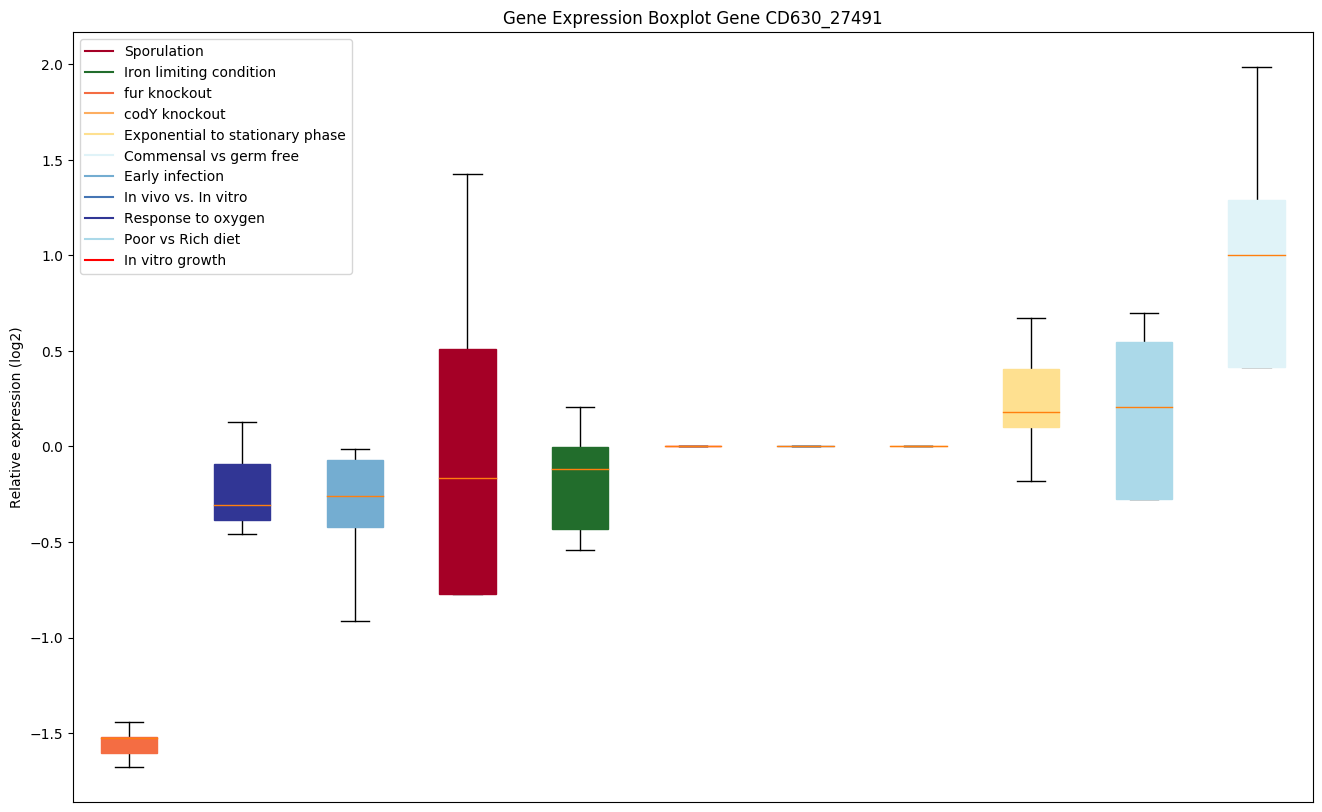 |
||||
| CD630_31730 | pgk | Phosphoglycerate kinase | Yes | Yes |
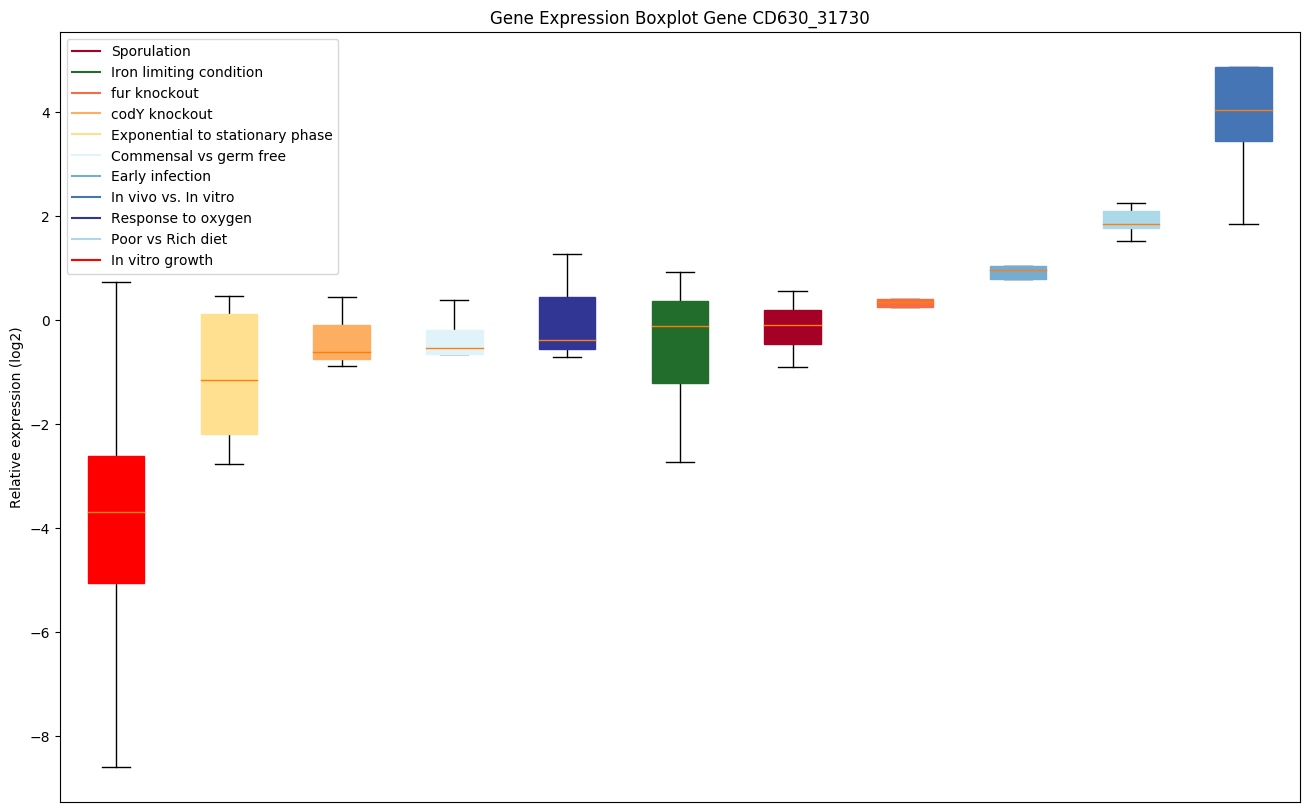 |
||
| CD630_07140 | Putative protein involved in protein-proteininteractions |
 |
|||||
| CD630_29750 | Putative CRISPR-associated Cas2 family protein | CRISPR (clustered regularly interspaced short palindromic repeat), is an adaptive immune system that provides protection against mobile genetic elements (viruses, transposable elements and conjugative plasmids). CRISPR clusters contain sequences complementary to antecedent mobile elements and target invading nucleic acids. CRISPR clusters are transcribed and processed into CRISPR RNA (crRNA). Functions as a ssRNA-specific endoribonuclease. Involved in the integration of spacer DNA into the CRISPR cassette. |
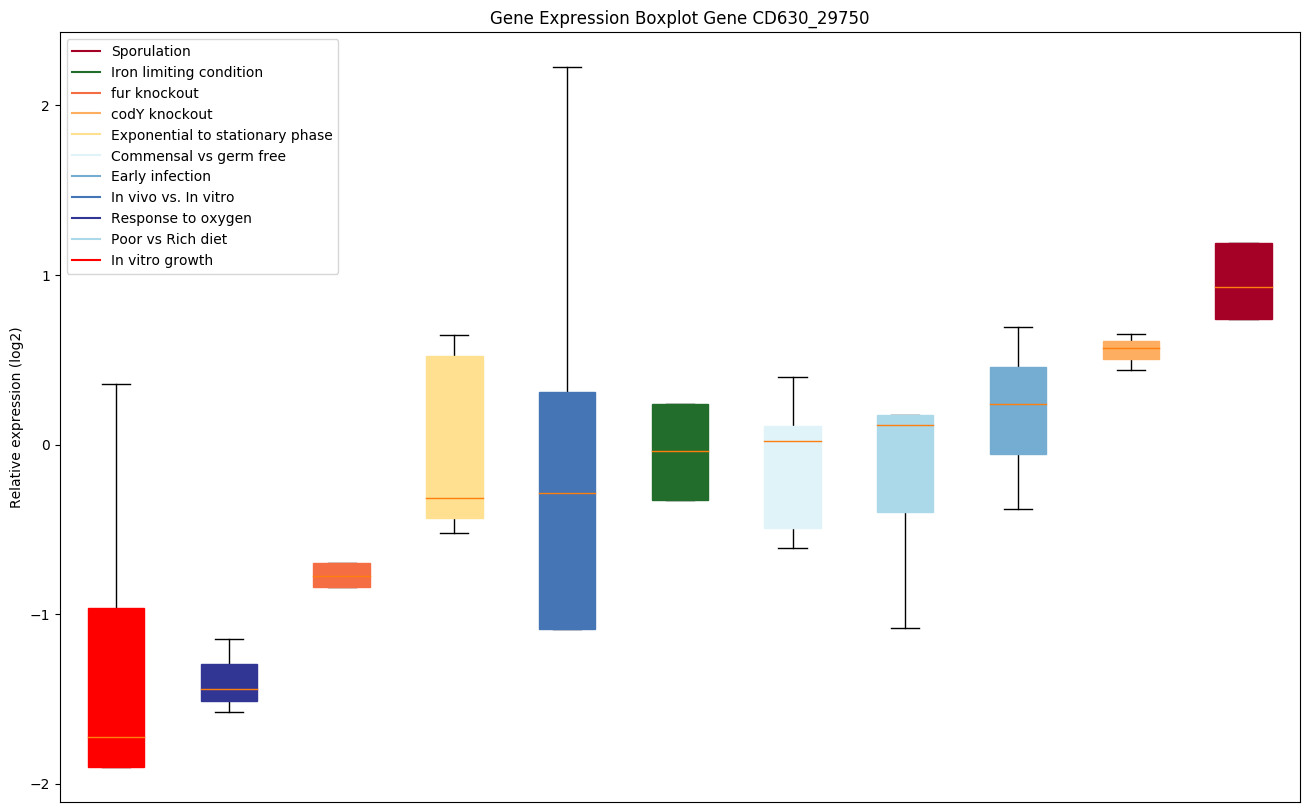 |
||||
| CD630_07121 | Conserved hypothetical protein |
 |
|||||
| CD630_31740 | gapA | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) |
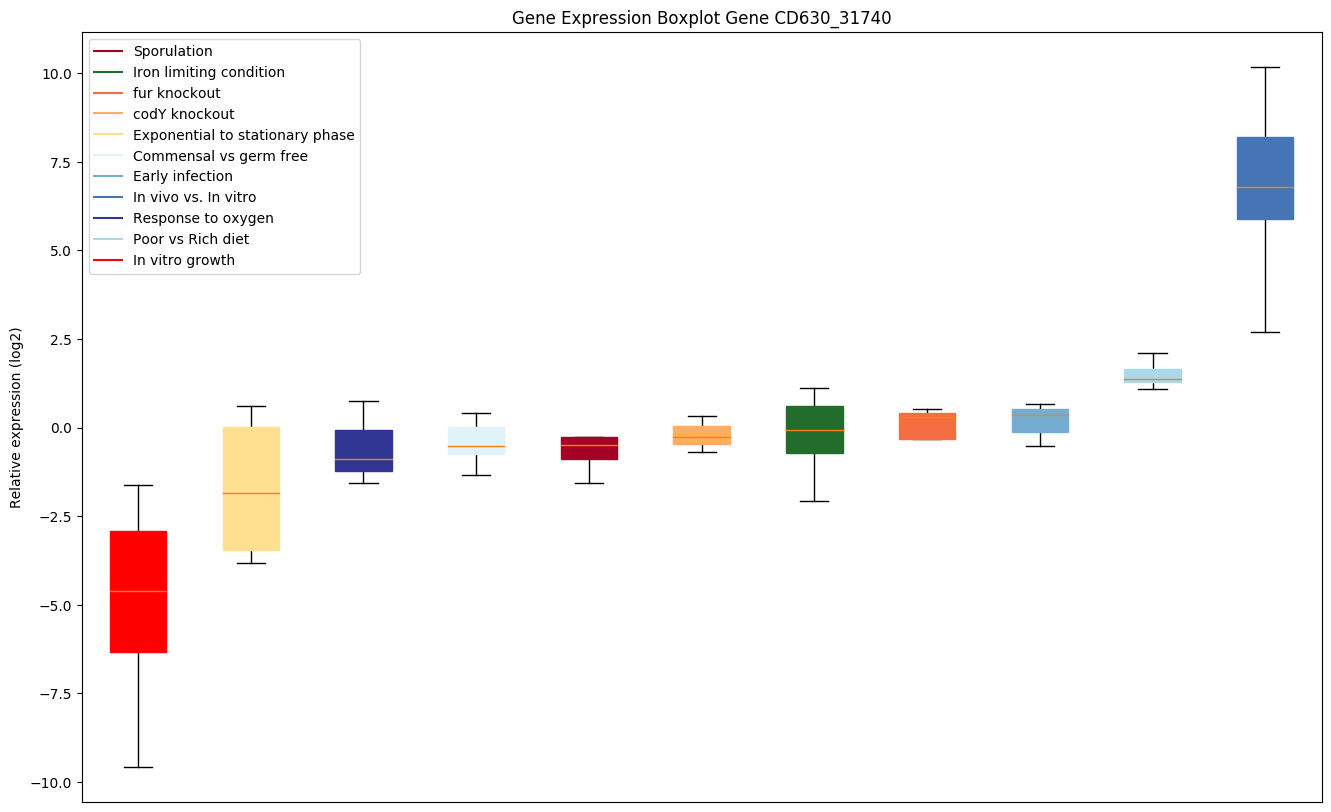 |
||||
| CD630_07130 | Putative radical SAM domain-containing protein |
 |
|||||
| CD630_29810 | Putative CRISPR-associated protein, CXXC-CXXC |
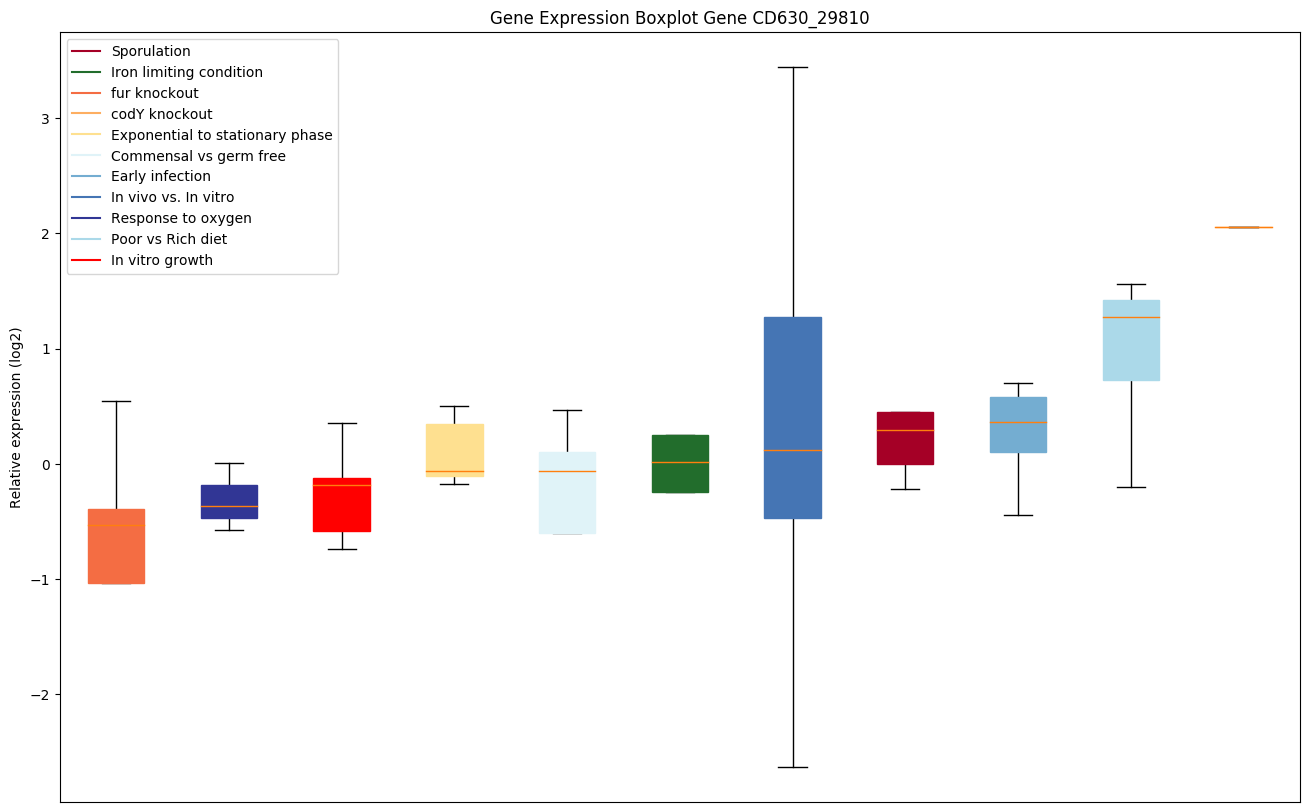 |
|||||
| CD630_20720 | Putative transposase |
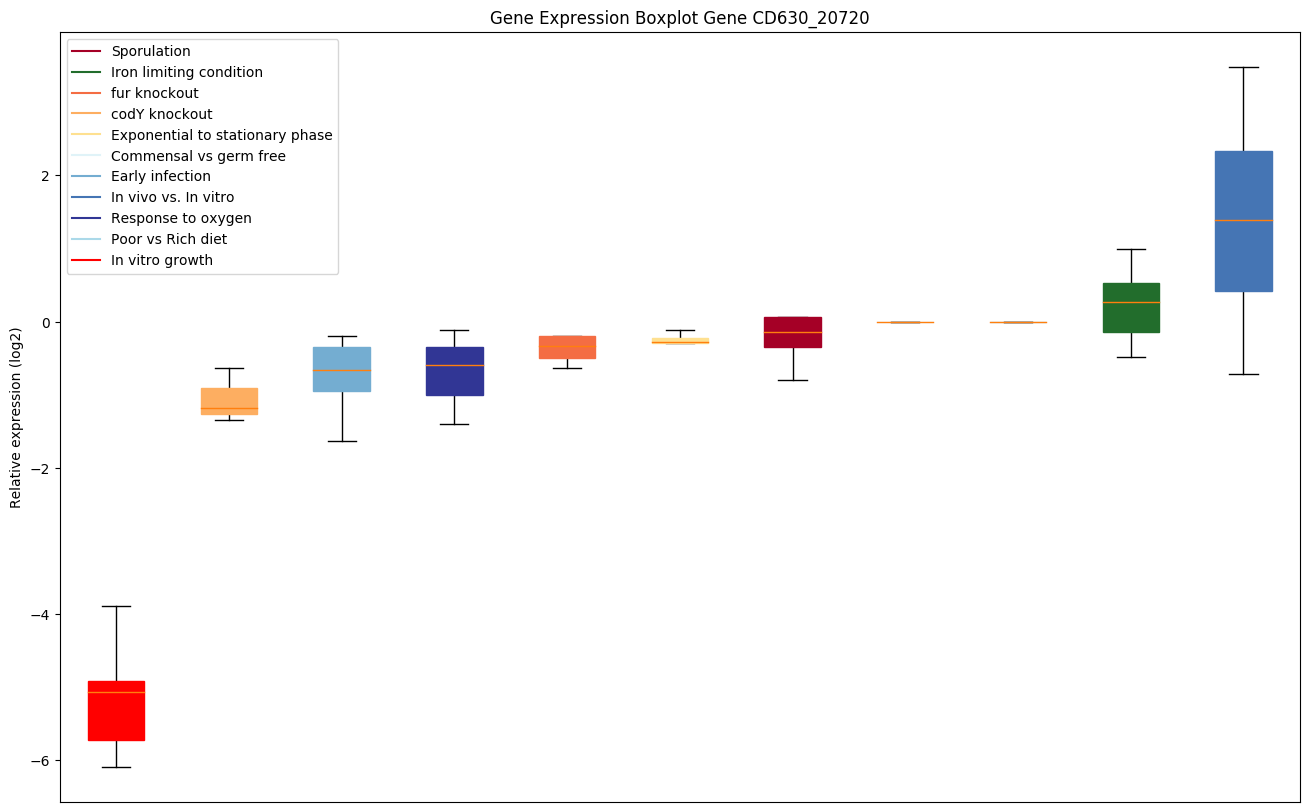 |
|||||
| CD630_31750 | cggR | Transcriptional regulator, SorC family |
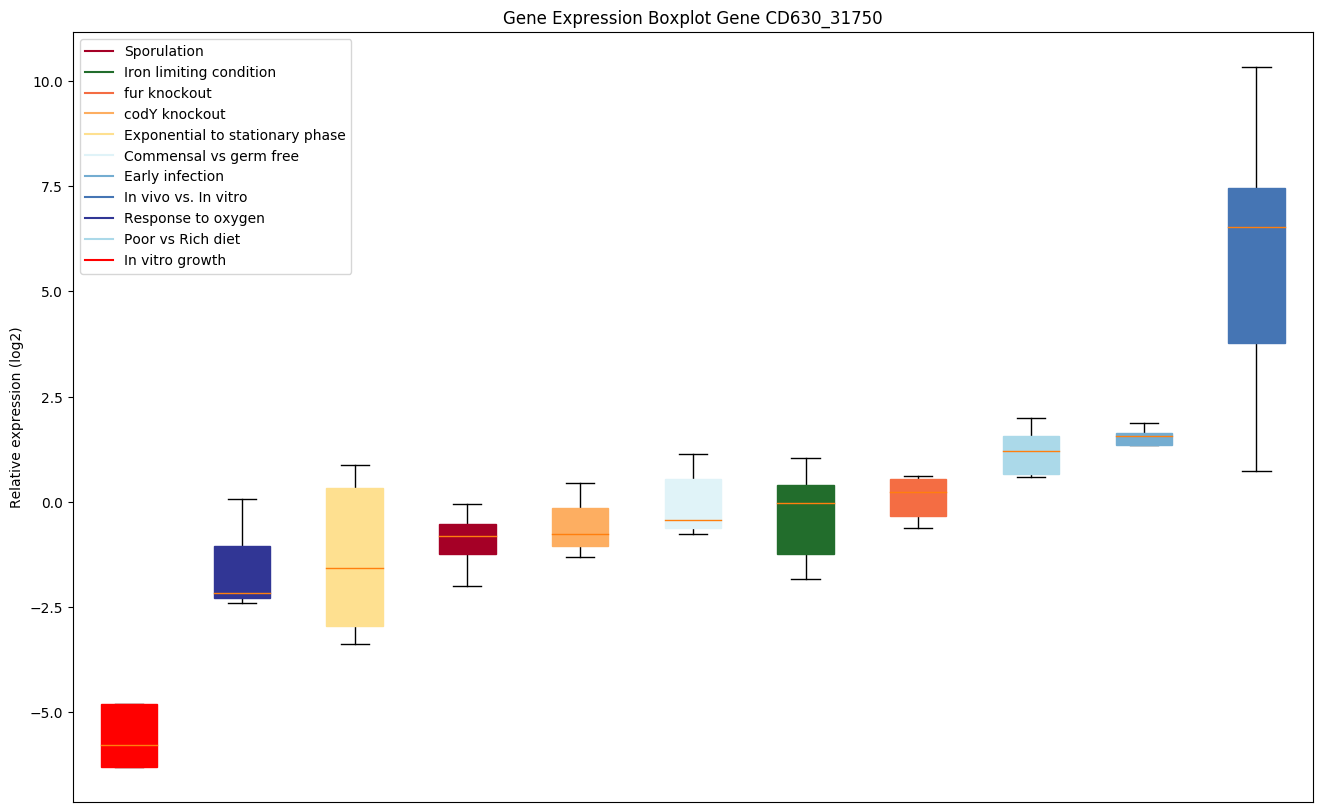 |
||||
| CD630_29800 | Putative CRISPR-associated autoregulator DevRfamily protein protein |
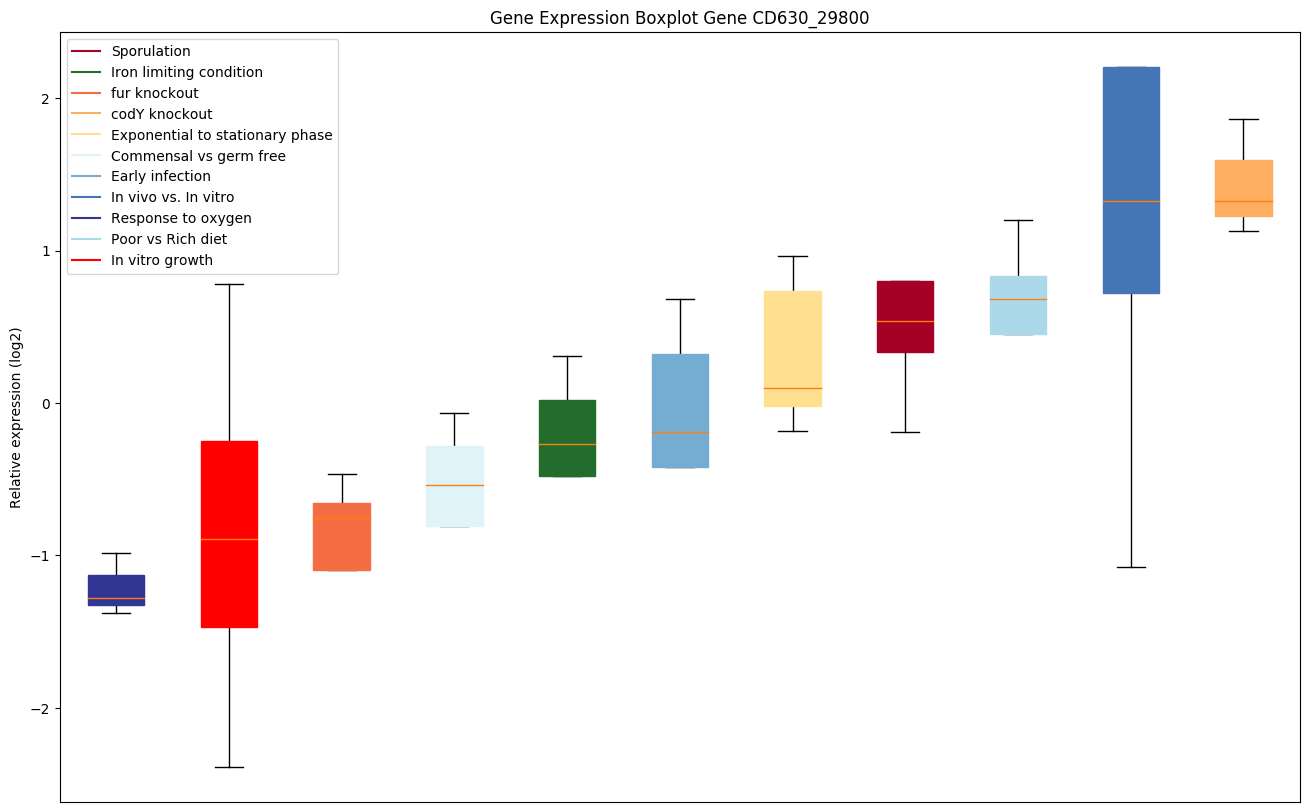 |