Module 374
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.57 | 20 | 64 | NA | NA |
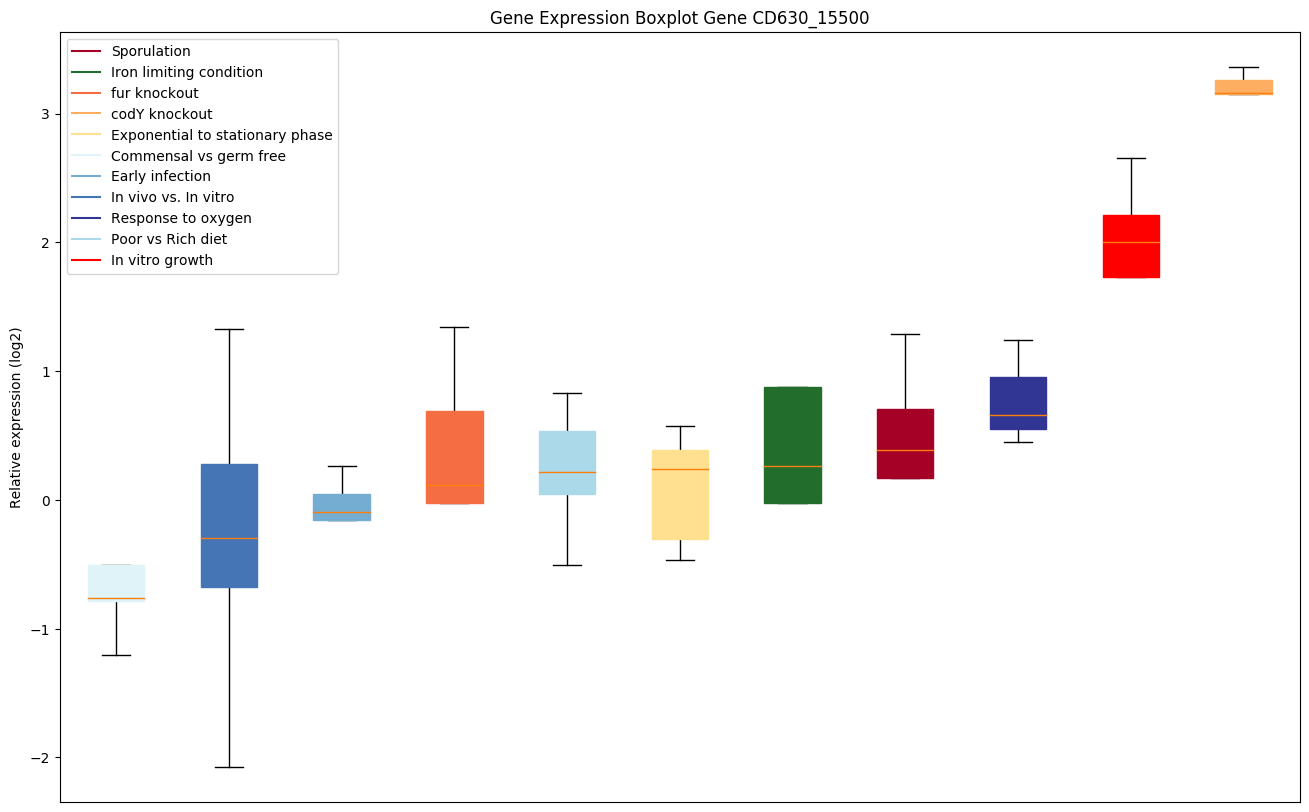
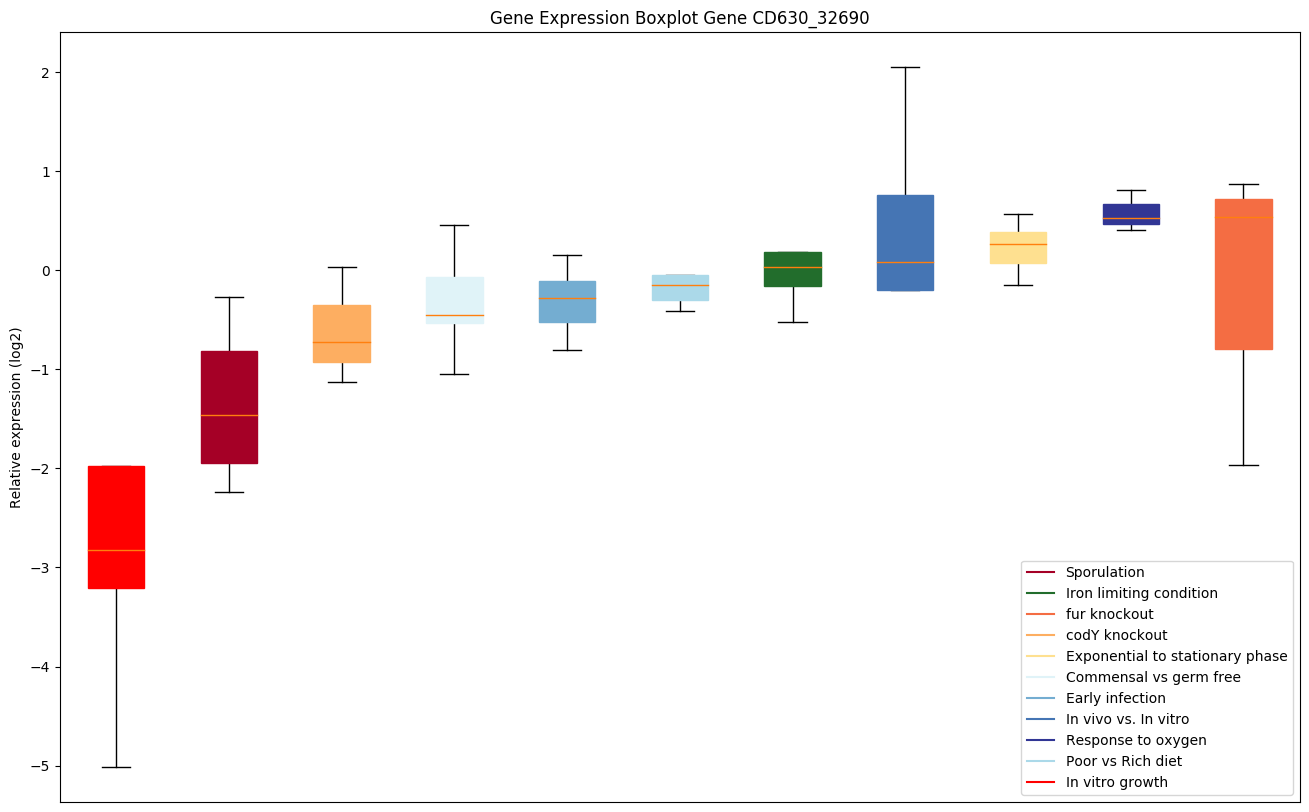
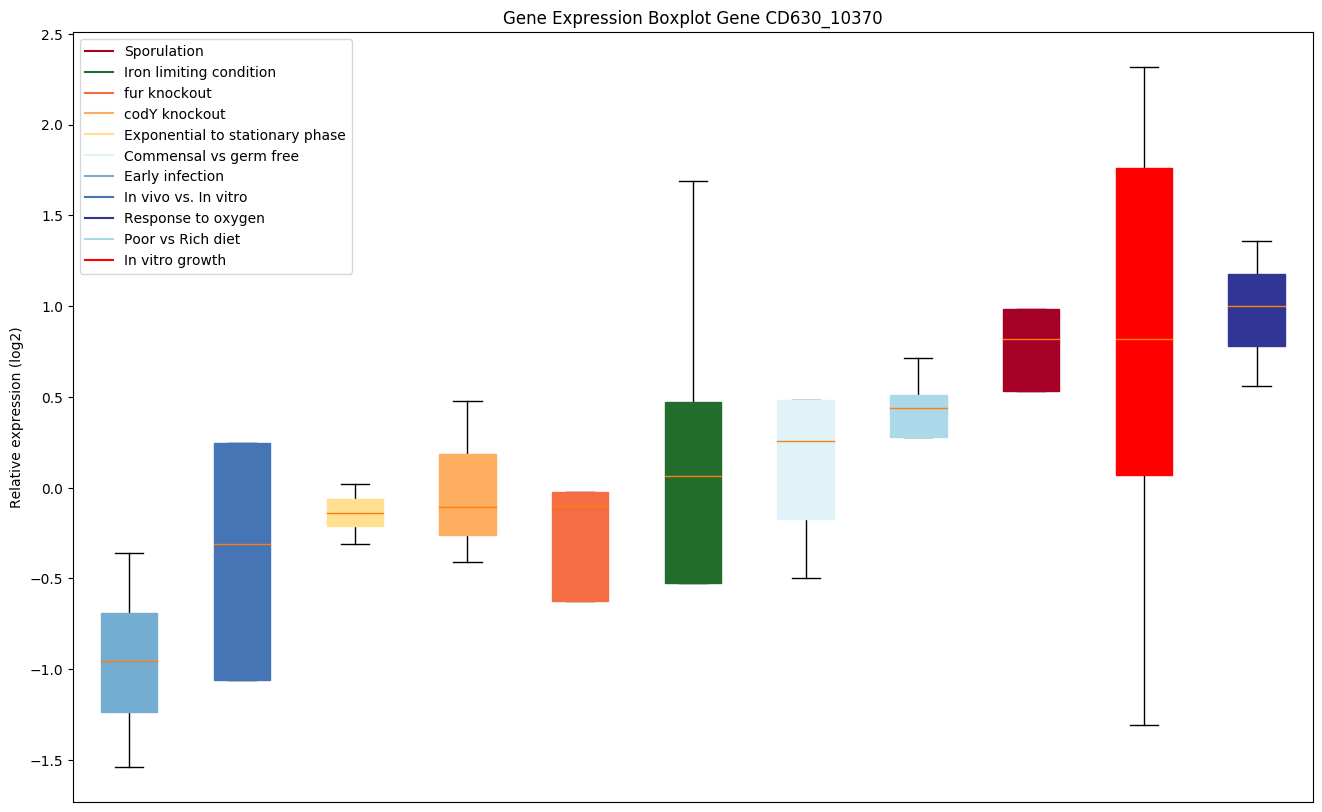
Bicluster expression profile
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
| TF | Module | Confidence Score | Beta |
|---|---|---|---|
|
Transcriptional regulator, HTH-type |
374 | 0.74 | 0.4785 |
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
| TF | Module | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
|
codY Transcriptional regulator, GTP-sensingpleiotropic repressor |
374 | 5 | 0.00 | 0.03 |
The module is significantly enriched with genes associated to the indicated functional terms
The enrichment was determined with hypergeometric test using the genome functional annotation compiled by Girinathan et al (2020).
| Module | Pathway | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
| 374 | Histidine Biosynthesis | 7 | 0.000000 | 0.000000 |
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_15500 | hisB | Imidazoleglycerol-phosphate dehydratase |
 |
||||
| CD630_32690 | Putative oligoendopeptidase F, M3B family |
 |
|||||
| CD630_10370 | Conserved hypothetical protein |
 |
|||||
| CD630_15530 | hisF | Imidazole glycerol phosphate synthase subunitHisF | IGPS catalyzes the conversion of PRFAR and glutamine to IGP, AICAR and glutamate. The HisF subunit catalyzes the cyclization activity that produces IGP and AICAR from PRFAR using the ammonia provided by the HisH subunit. |
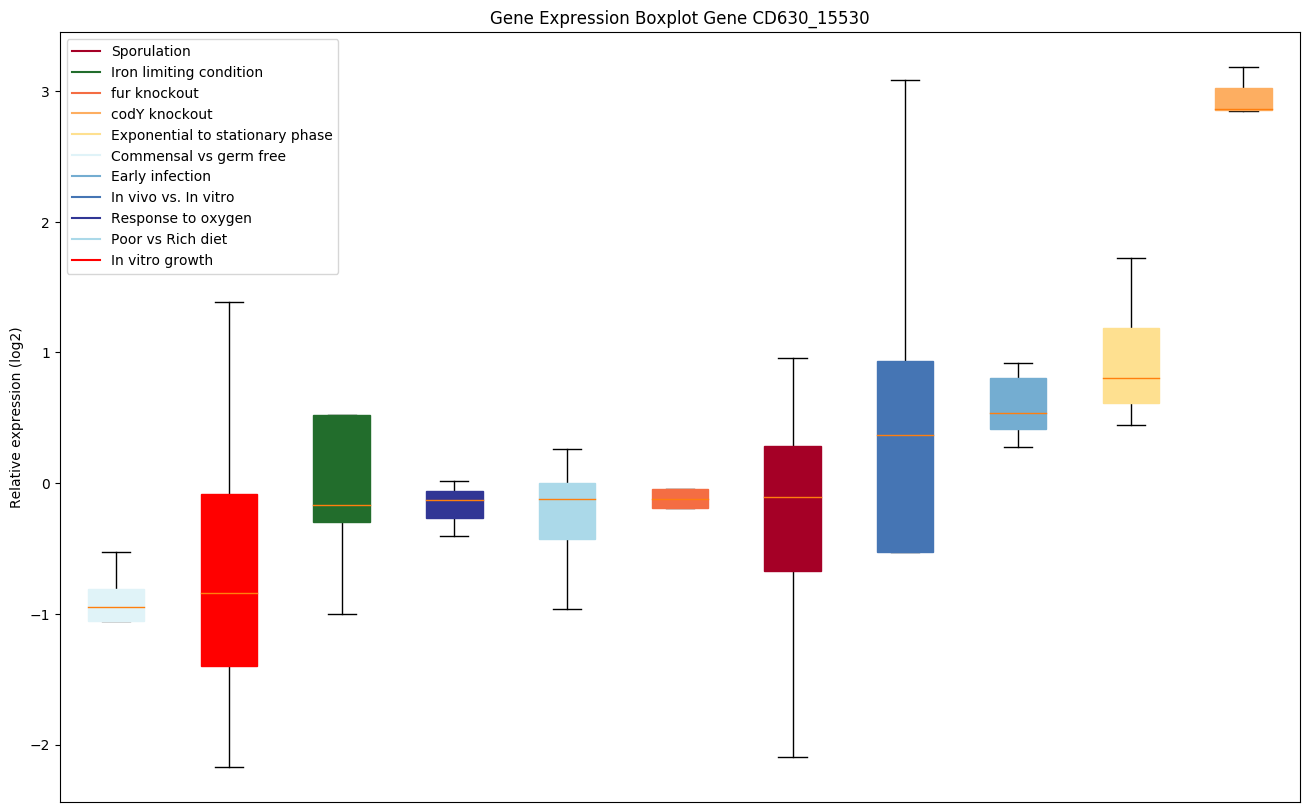 |
|||
| CD630_15480 | hisG | ATP phosphoribosyltransferase (ATP-PRTase)(ATP-PRT) | Catalyzes the condensation of ATP and 5-phosphoribose 1-diphosphate to form N'-(5'-phosphoribosyl)-ATP (PR-ATP). Has a crucial role in the pathway because the rate of histidine biosynthesis seems to be controlled primarily by regulation of HisG enzymatic activity. |
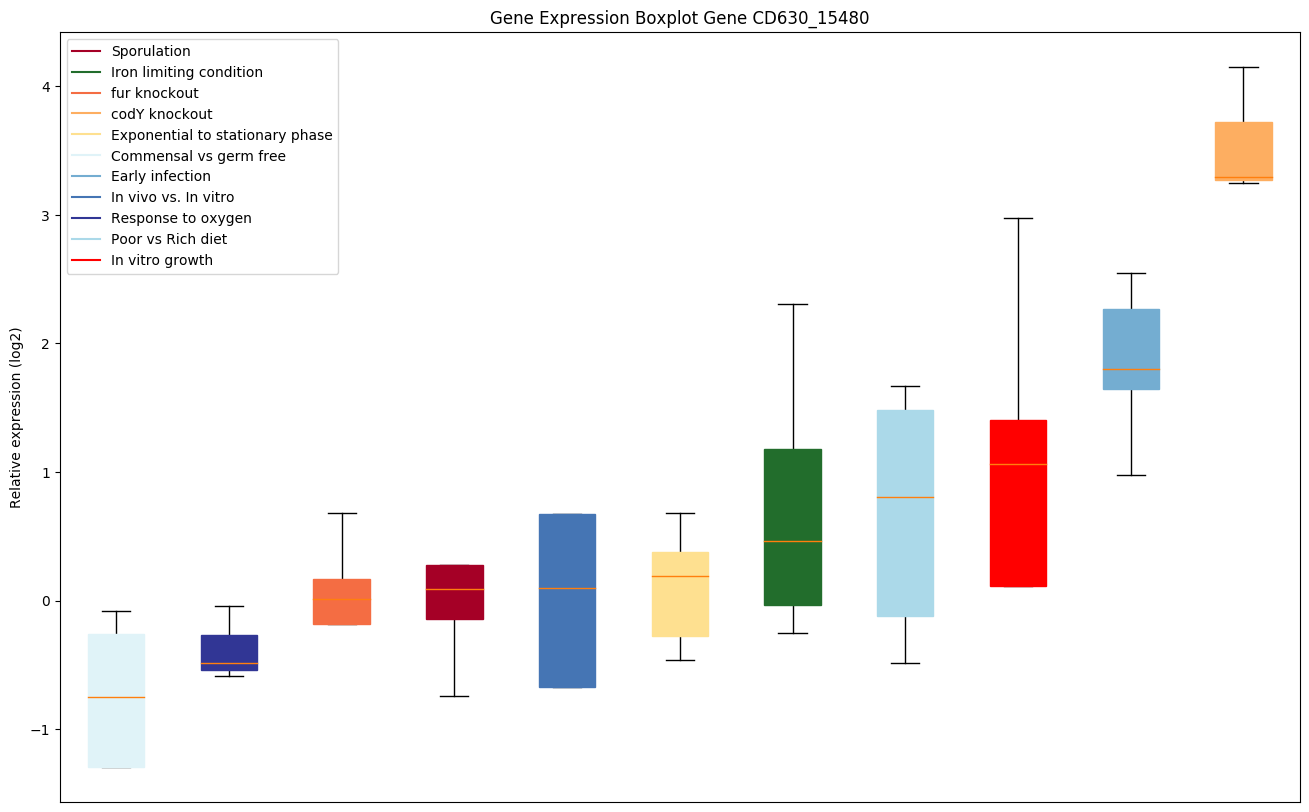 |
|||
| CD630_10430 | sbcC | Nuclease SbcCD subunit C |
 |
||||
| CD630_15520 | hisA | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase |
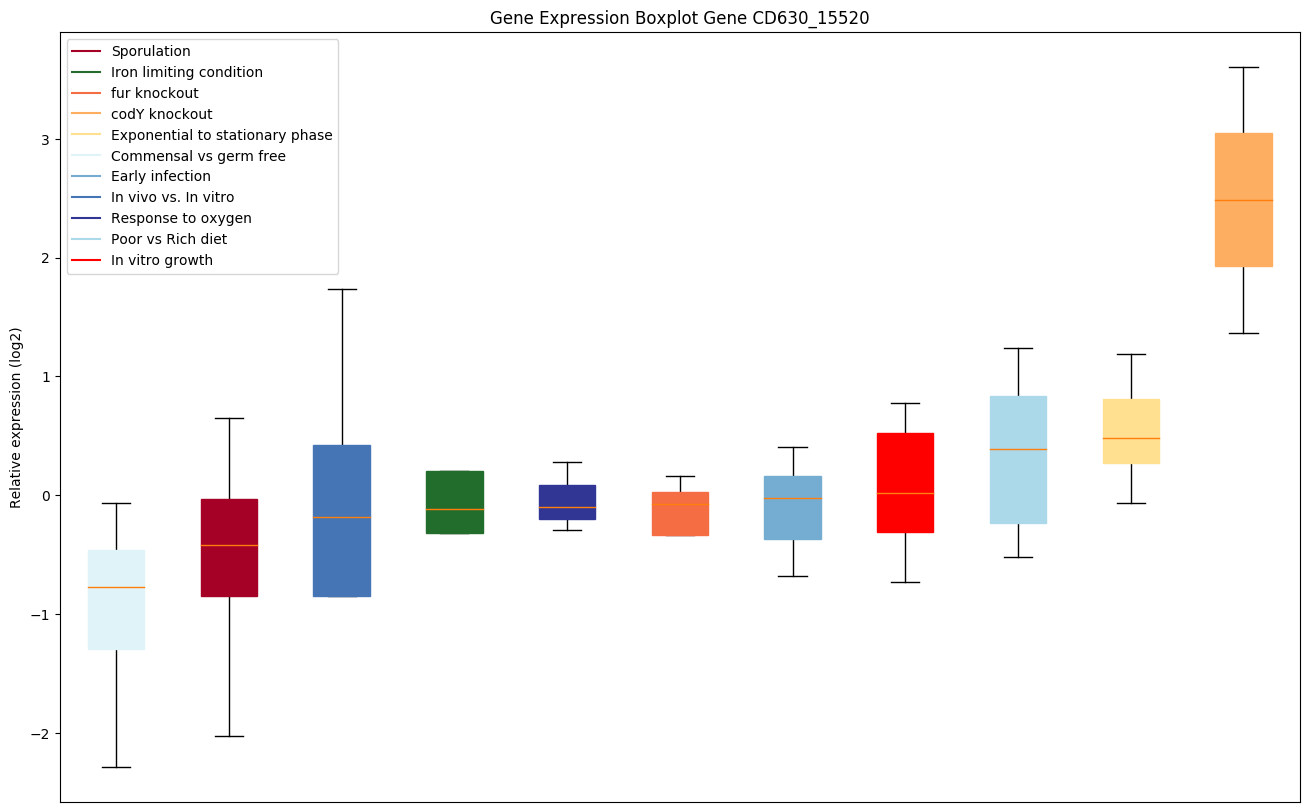 |
||||
| CD630_32700 | Putative magnesium transport ATPase, MgtC/SapBfamily |
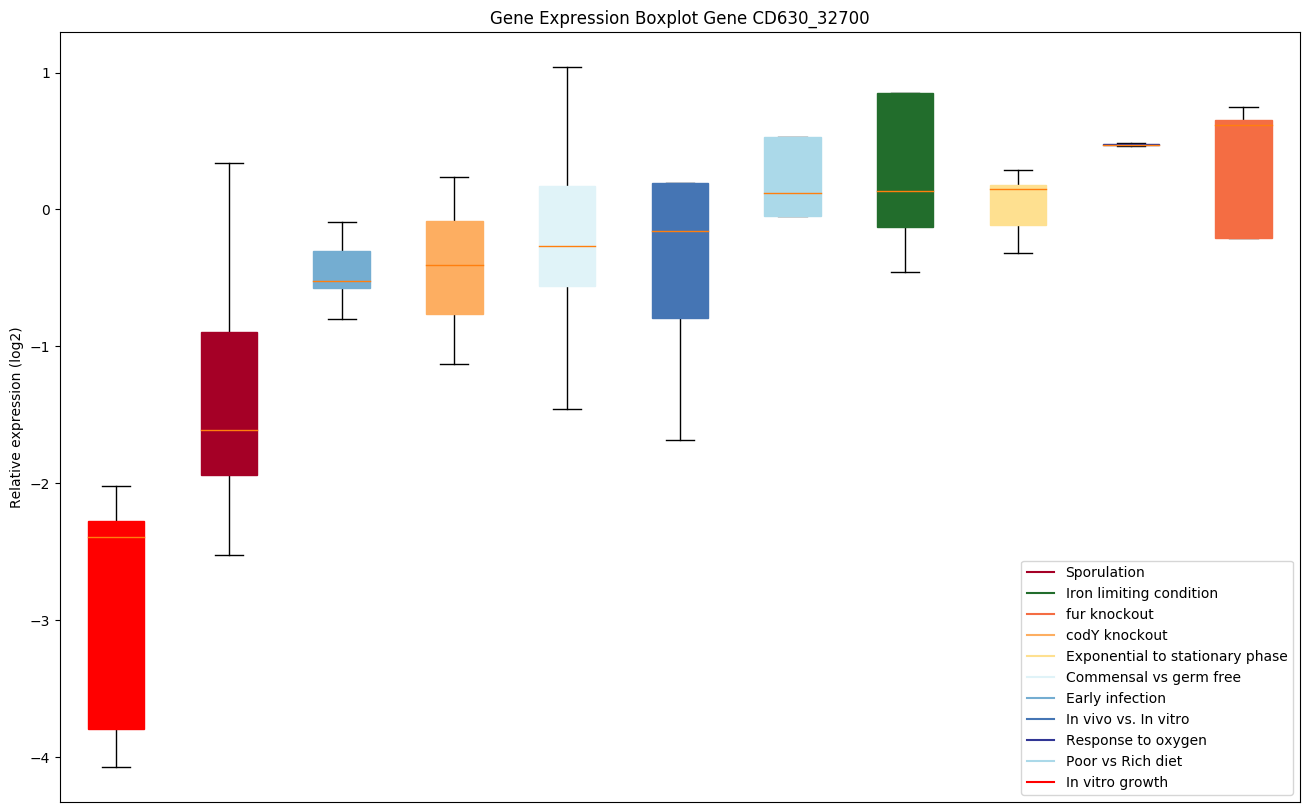 |
|||||
| CD630_15470 | hisZ | ATP phosphoribosyltransferase regulatory subunit |
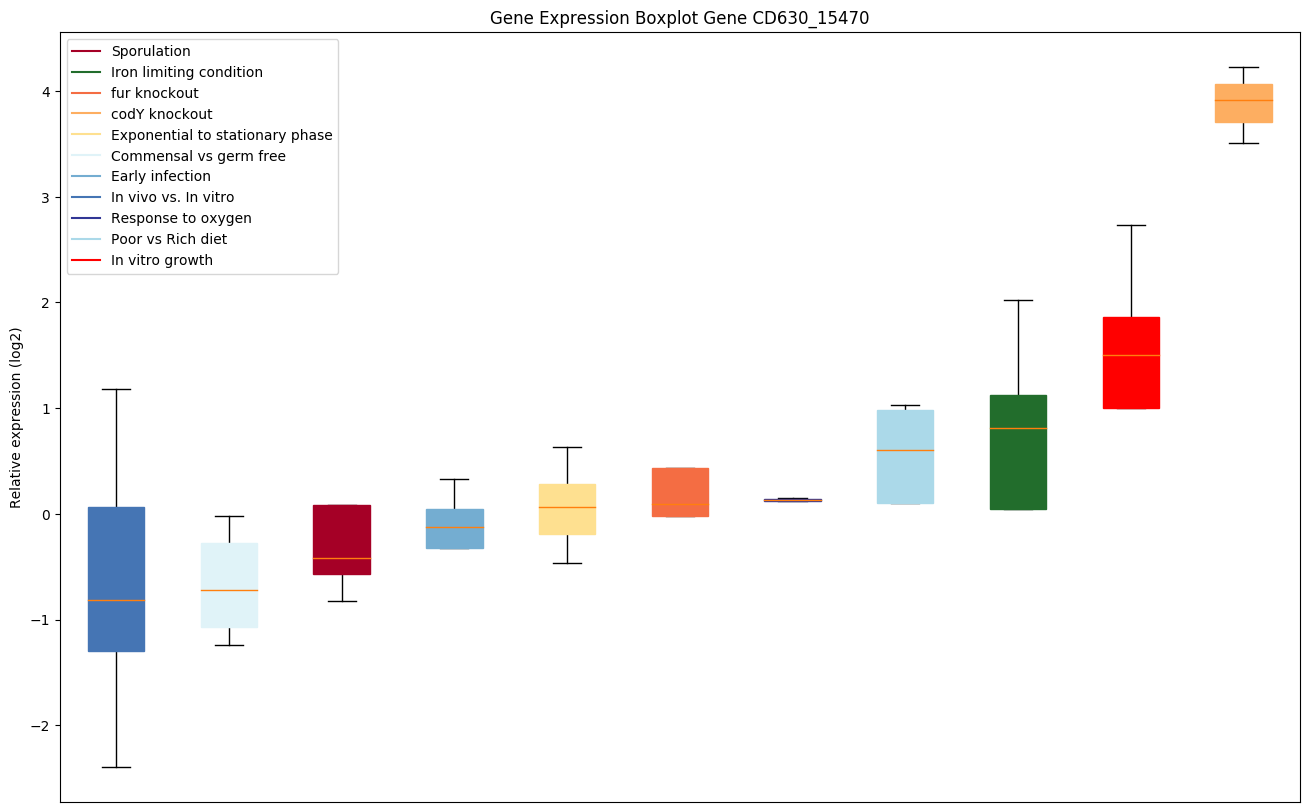 |
||||
| CD630_15490 | hisC | Histidinol-phosphate aminotransferase (Imidazoleacetol-phosphate transaminase) |
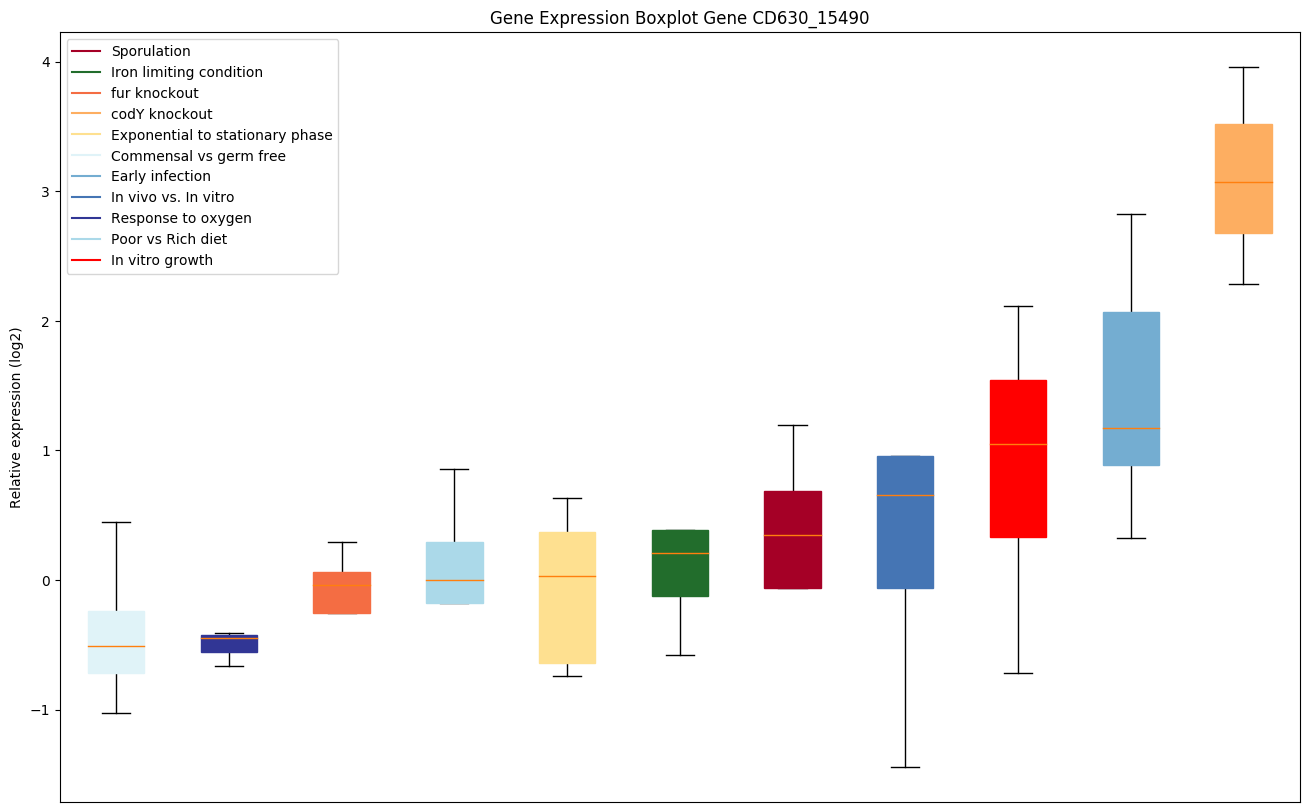 |
||||
| CD630_10410 | addA | ATP-dependent nuclease subunit A (ATP-dependenthelicase addA) | The heterodimer acts as both an ATP-dependent DNA helicase and an ATP-dependent, dual-direction single-stranded exonuclease. Recognizes the chi site generating a DNA molecule suitable for the initiation of homologous recombination. The AddA nuclease domain is required for chi fragment generation; this subunit has the helicase and 3' -> 5' nuclease activities. |
 |
|||
| CD630_21430 | Transcriptional regulator, HTH-type |
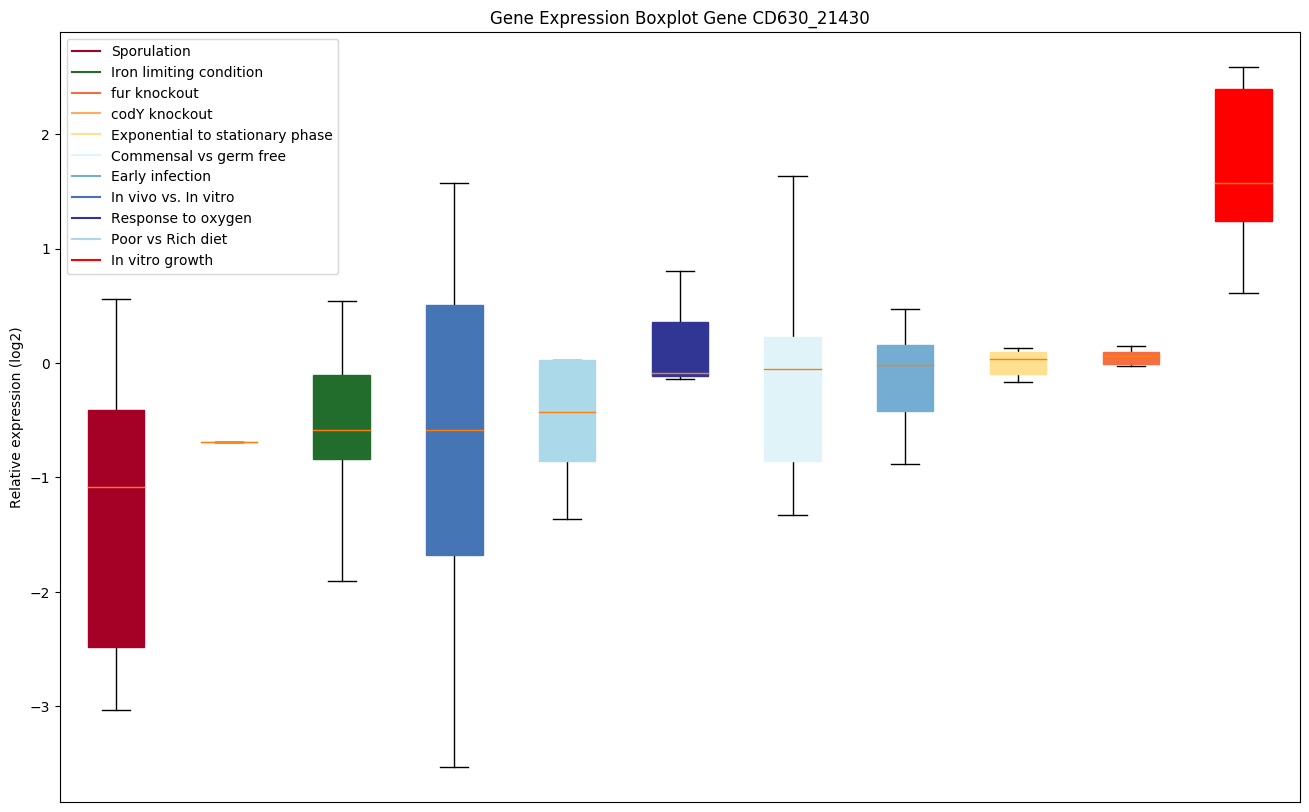 |
|||||
| CD630_10400 | addB | ATP-dependent nuclease subunit B (ATP-dependenthelicase addB), Superfamily 1 UvrD-family | The heterodimer acts as both an ATP-dependent DNA helicase and an ATP-dependent, dual-direction single-stranded exonuclease. Recognizes the chi site generating a DNA molecule suitable for the initiation of homologous recombination. The AddB nuclease domain is not required for chi fragment generation; this subunit has 5' -> 3' nuclease activity. | Yes |
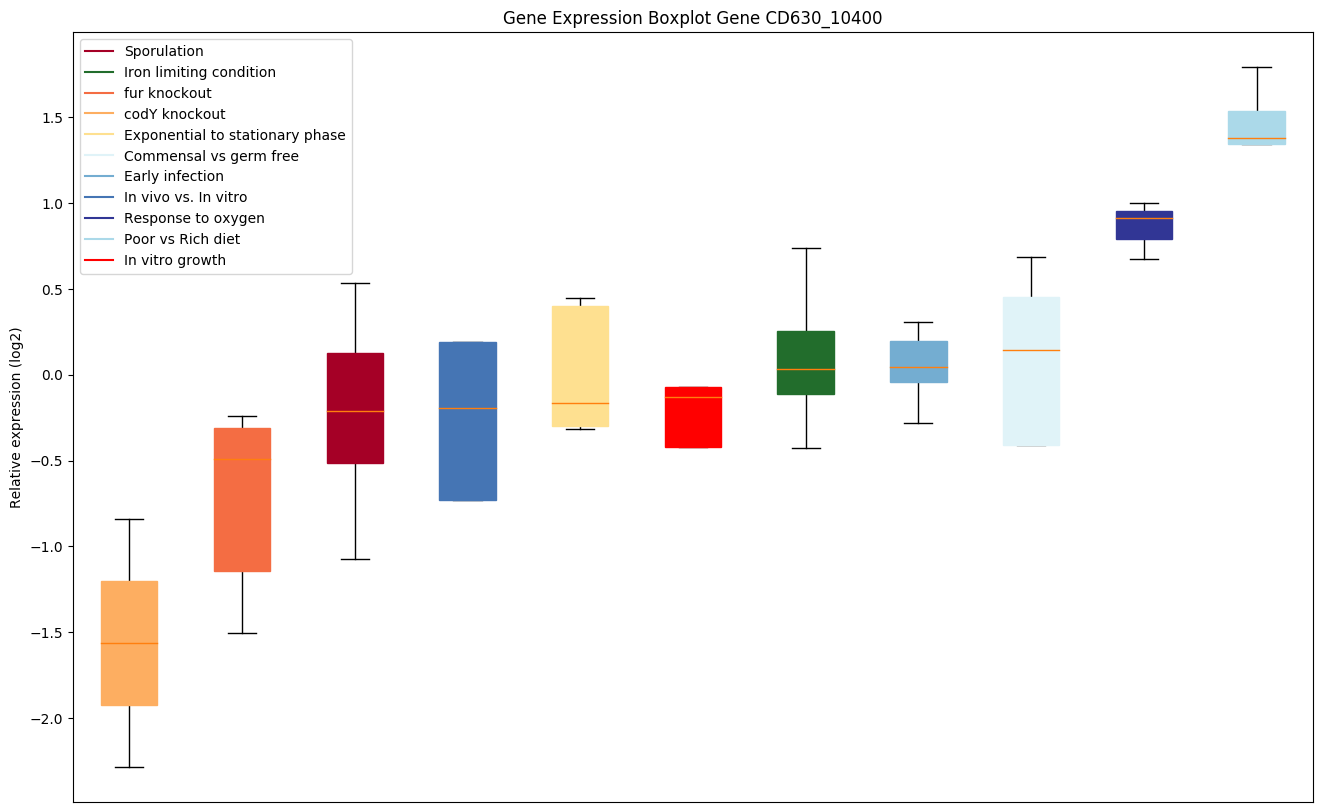 |
||
| CD630_10380 | Putative helicase, superfamily 1, UvrD-family |
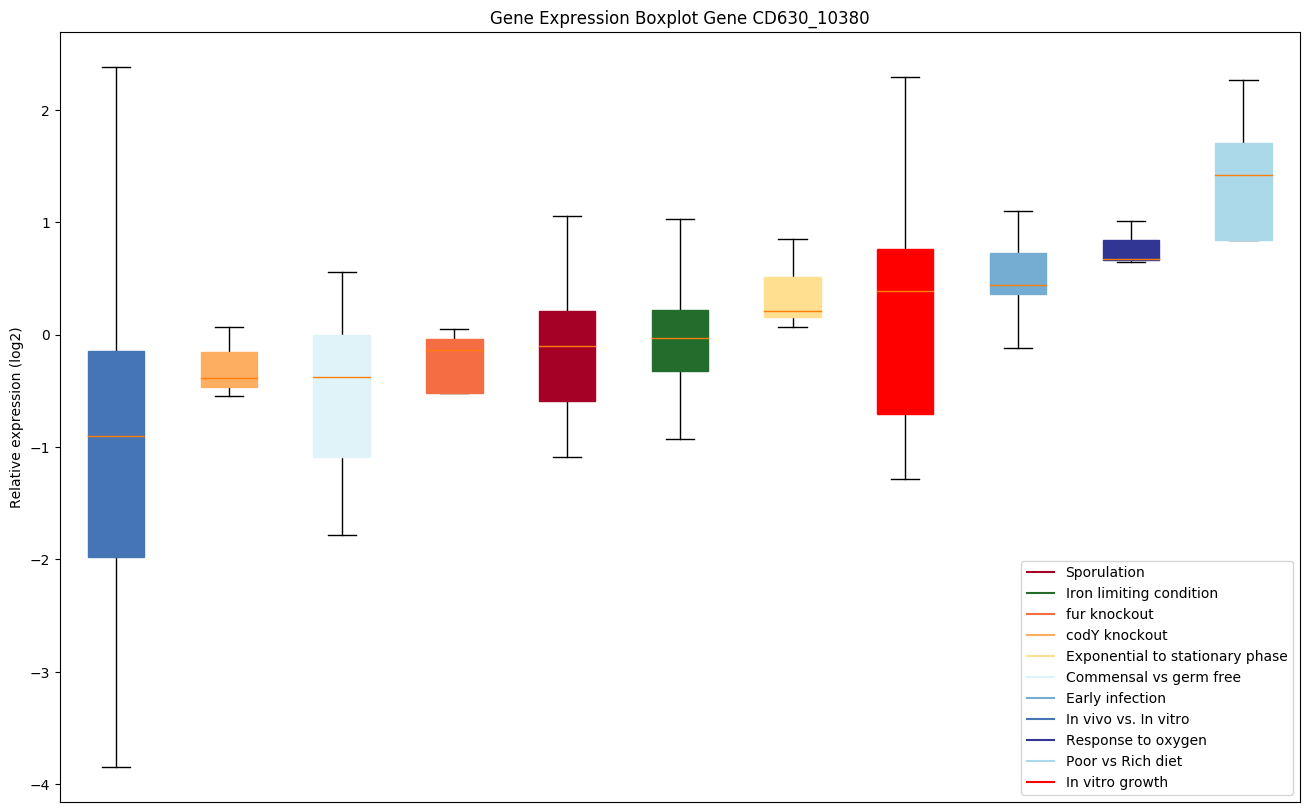 |
|||||
| CD630_10420 | sbcD | Nuclease SbcCD subunit D | SbcCD cleaves DNA hairpin structures. These structures can inhibit DNA replication and are intermediates in certain DNA recombination reactions. The complex acts as a 3'->5' double strand exonuclease that can open hairpins. It also has a 5' single-strand endonuclease activity. |
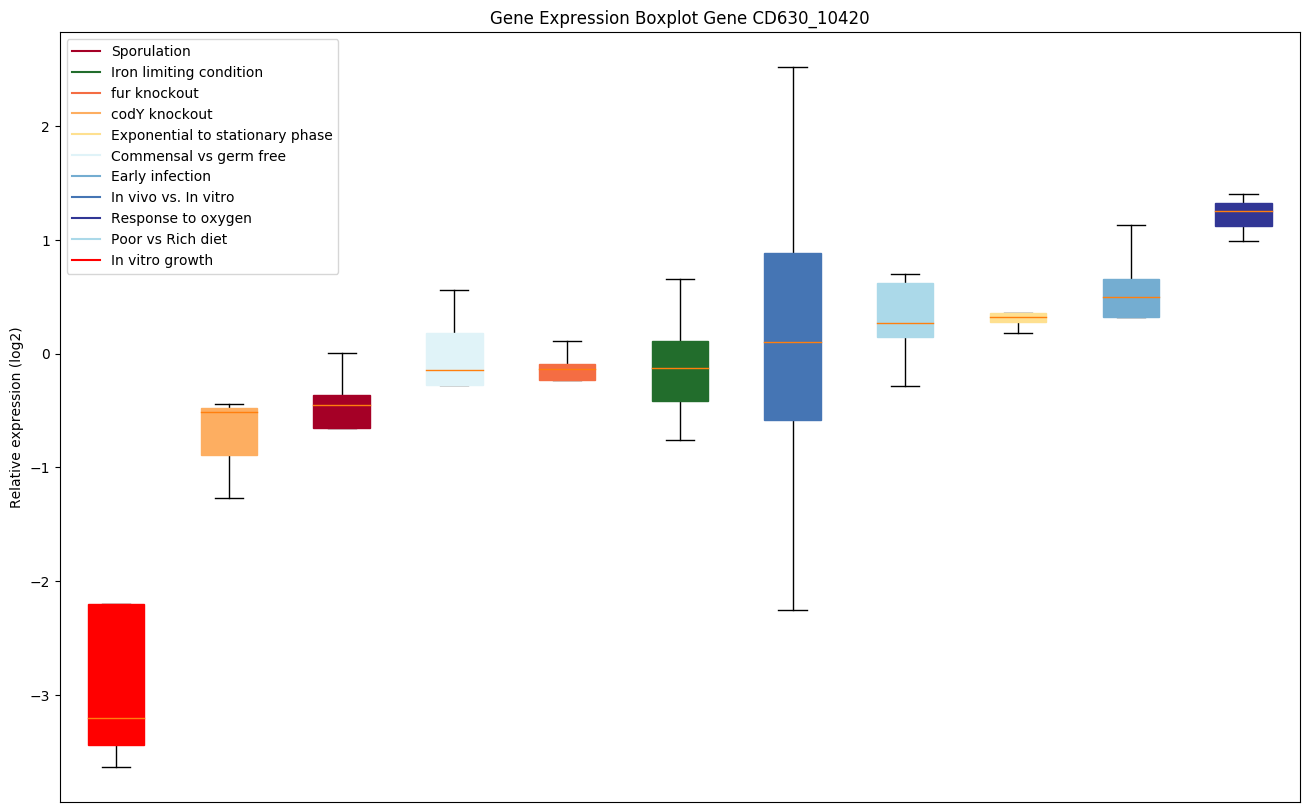 |
|||
| CD630_10390 | Conserved hypothetical protein |
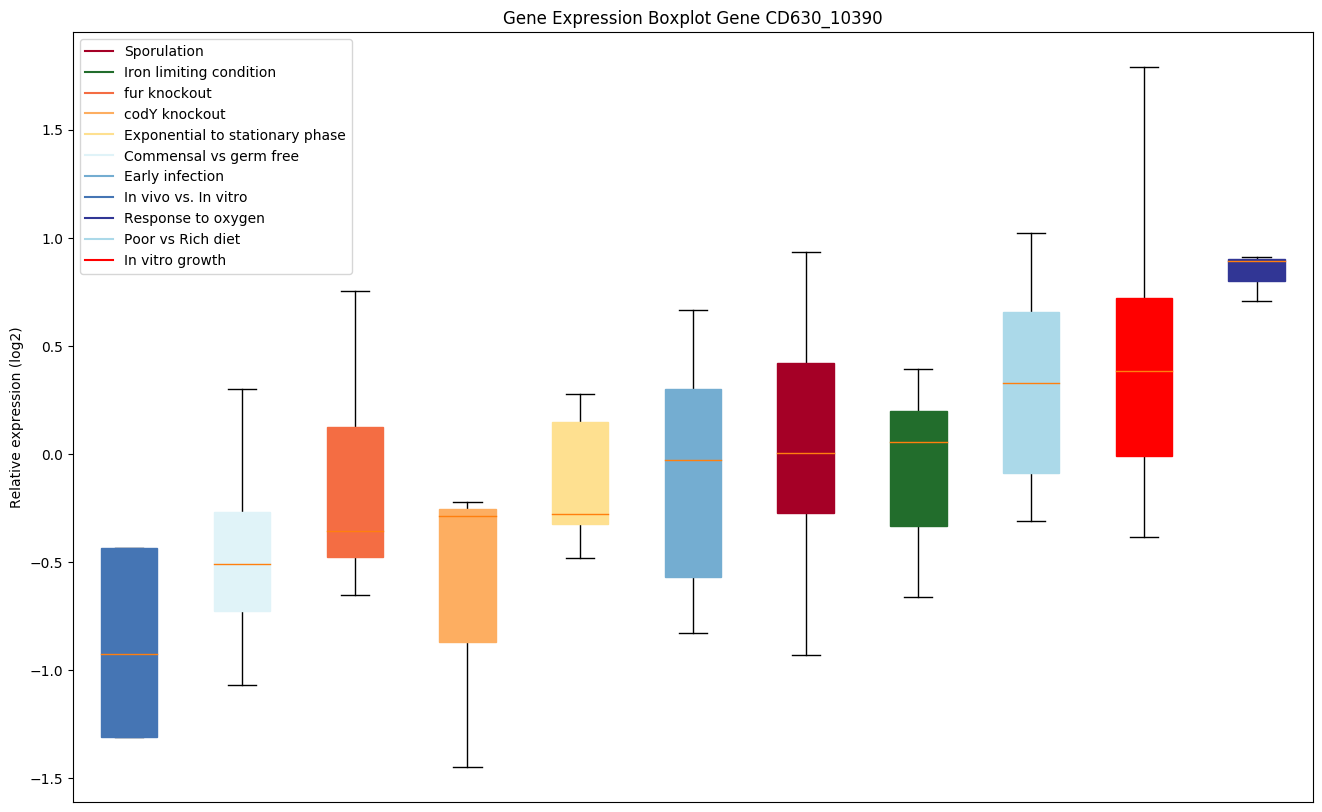 |
|||||
| CD630_15540 | hisI | Histidine biosynthesis bifunctional protein HisIE[Includes: Phosphoribosyl-AMP cyclohydrolase ;Phosphoribosyl-ATP pyrophosphatase] |
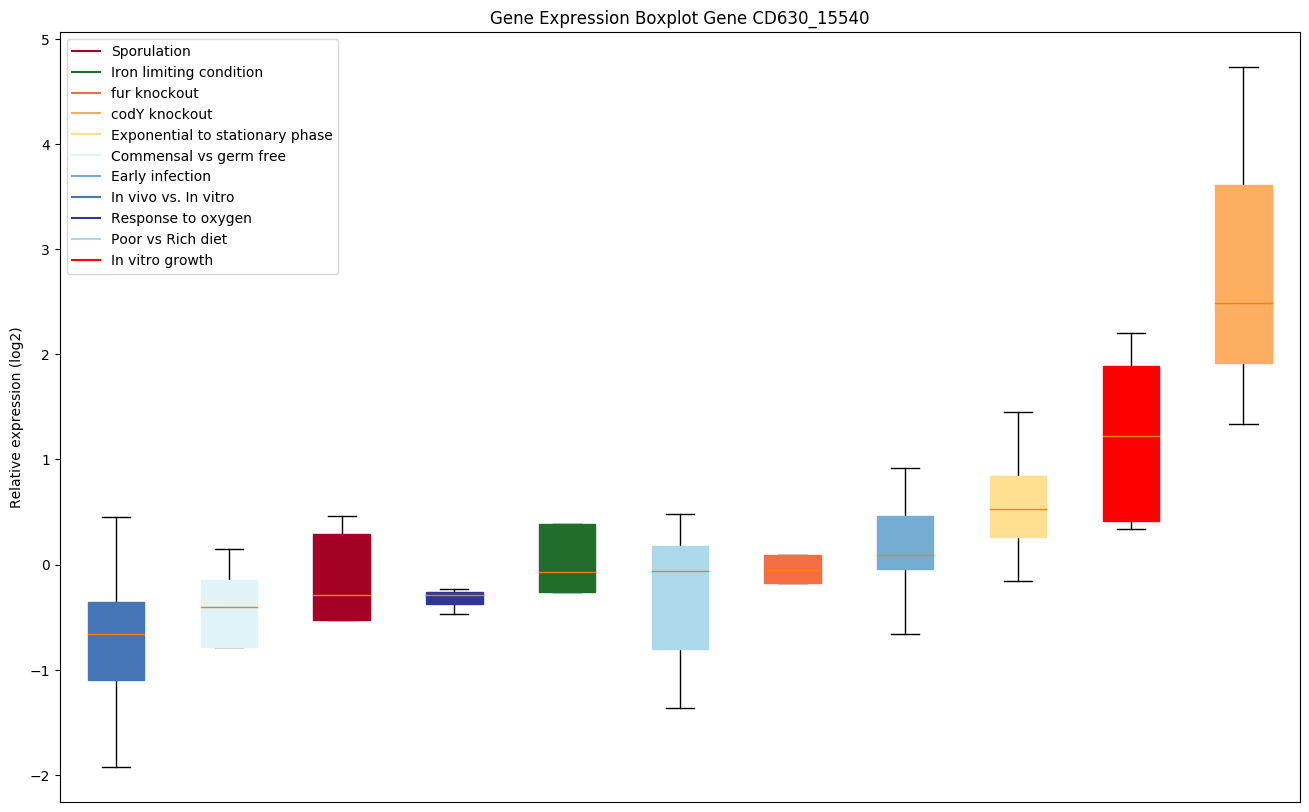 |
||||
| CD630_15510 | hisH | Imidazole glycerol phosphate synthase subunitHisH | IGPS catalyzes the conversion of PRFAR and glutamine to IGP, AICAR and glutamate. The HisH subunit catalyzes the hydrolysis of glutamine to glutamate and ammonia as part of the synthesis of IGP and AICAR. The resulting ammonia molecule is channeled to the active site of HisF. |
 |
|||
| CD630_21440 | Putative membrane protein | Probably functions as a manganese efflux pump. |
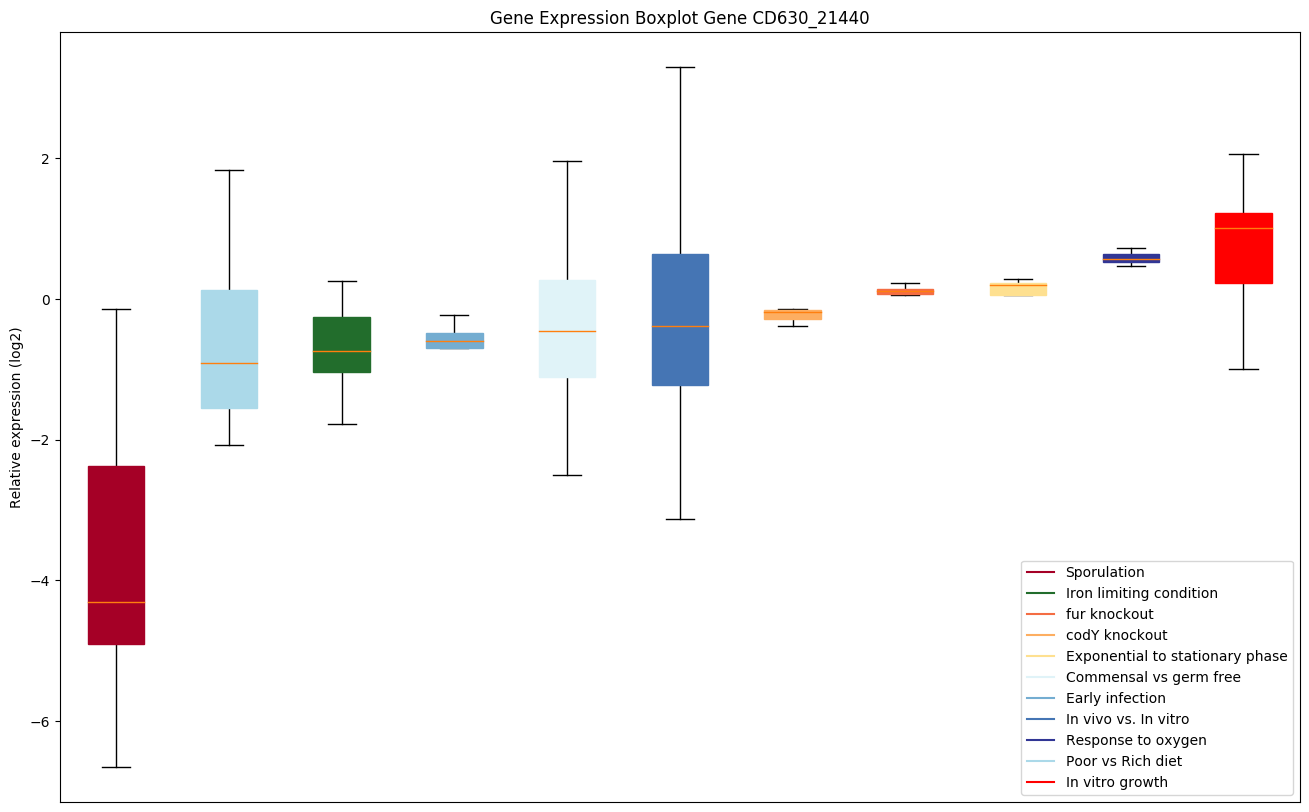 |
||||
| CD630_20020 | Transcriptional regulator, HTH-type |
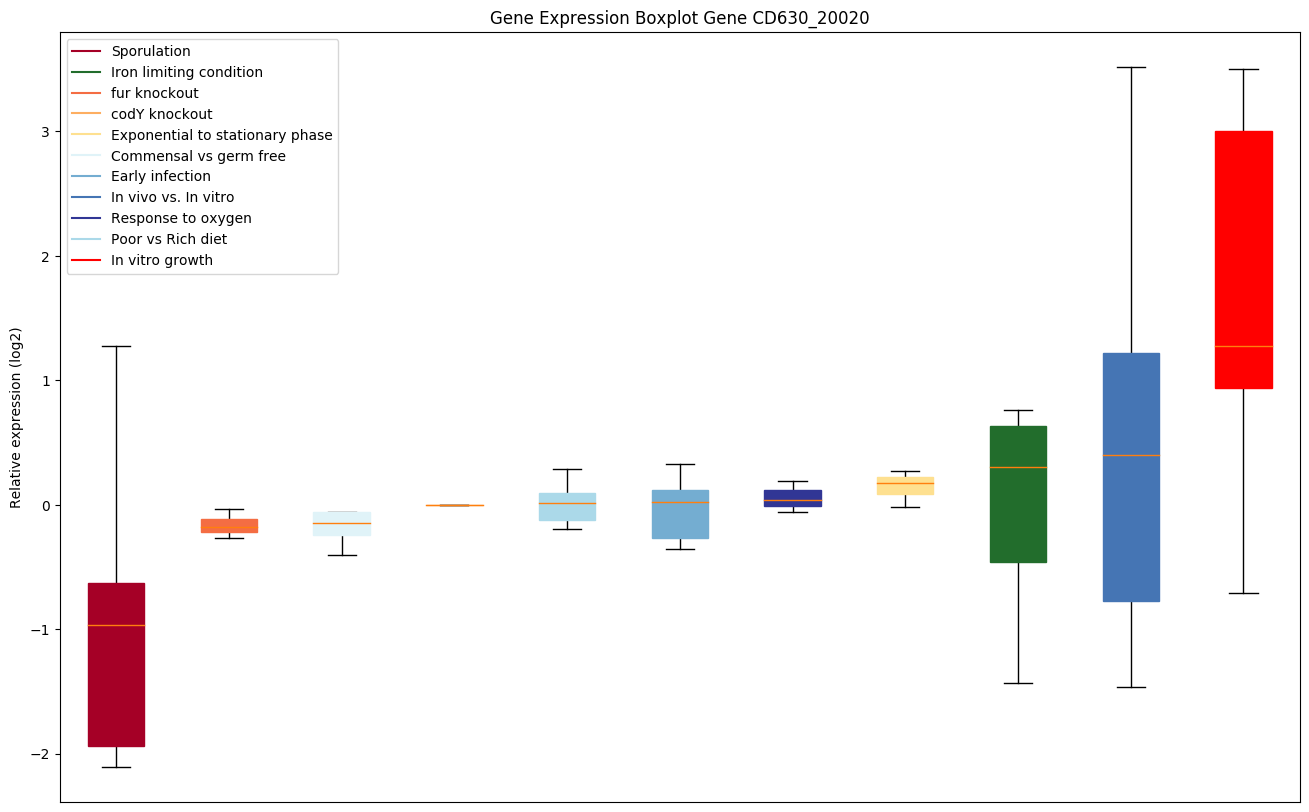 |