Module 50
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.61 | 42 | 64 | NA | NA |
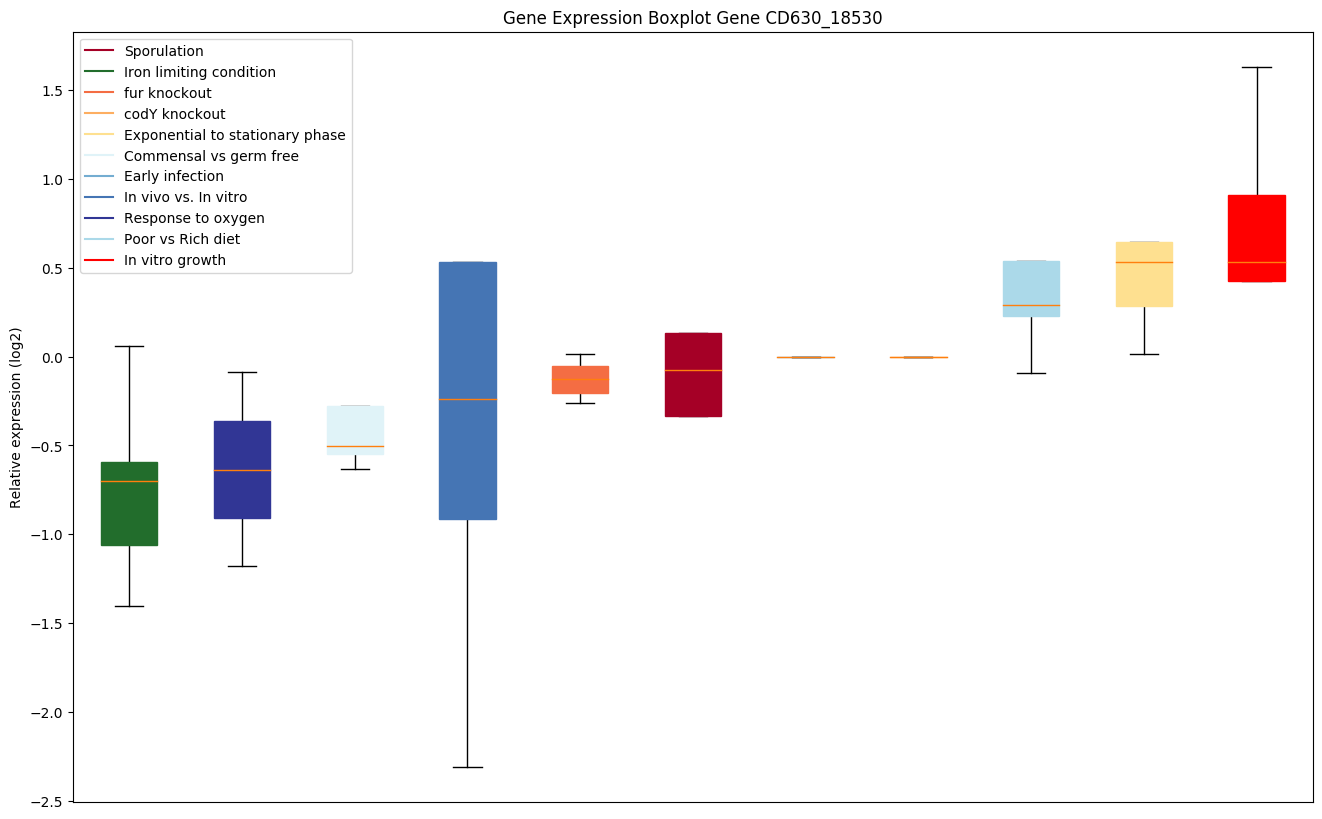
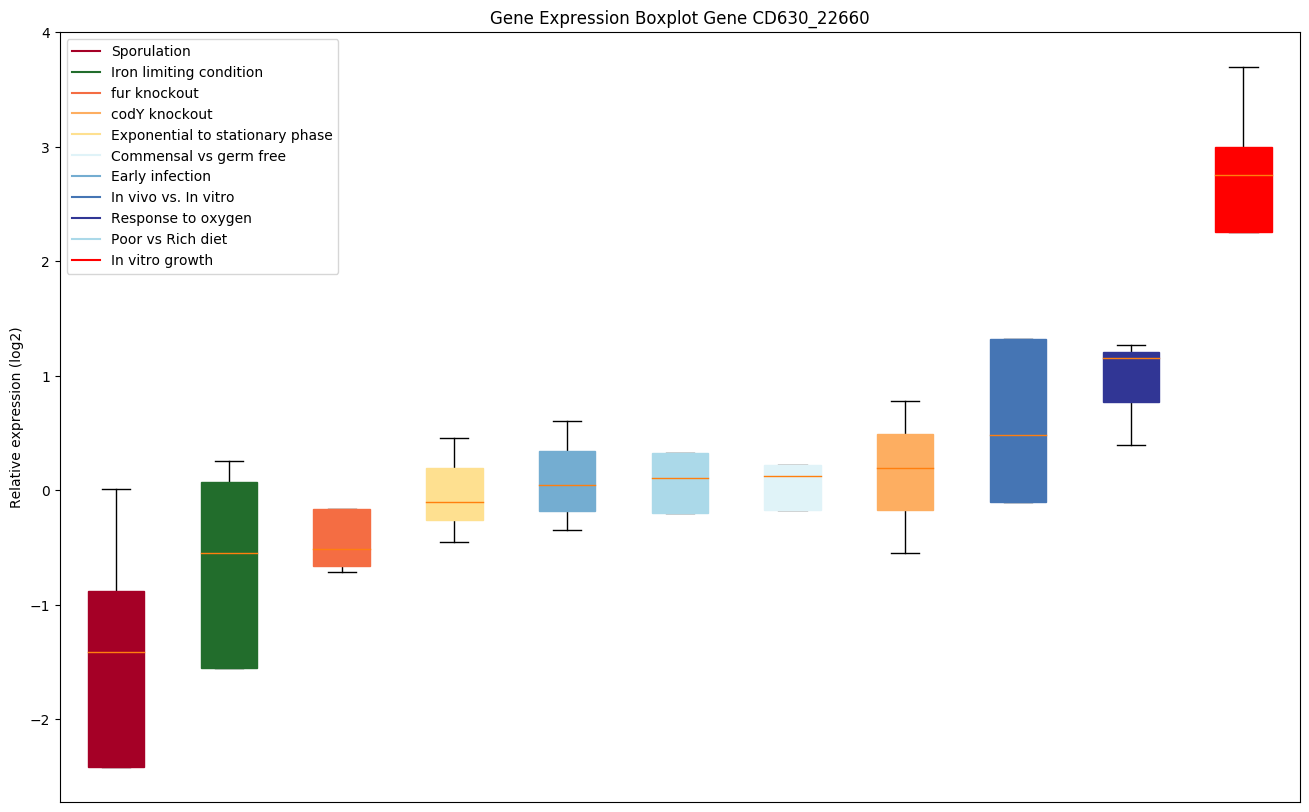
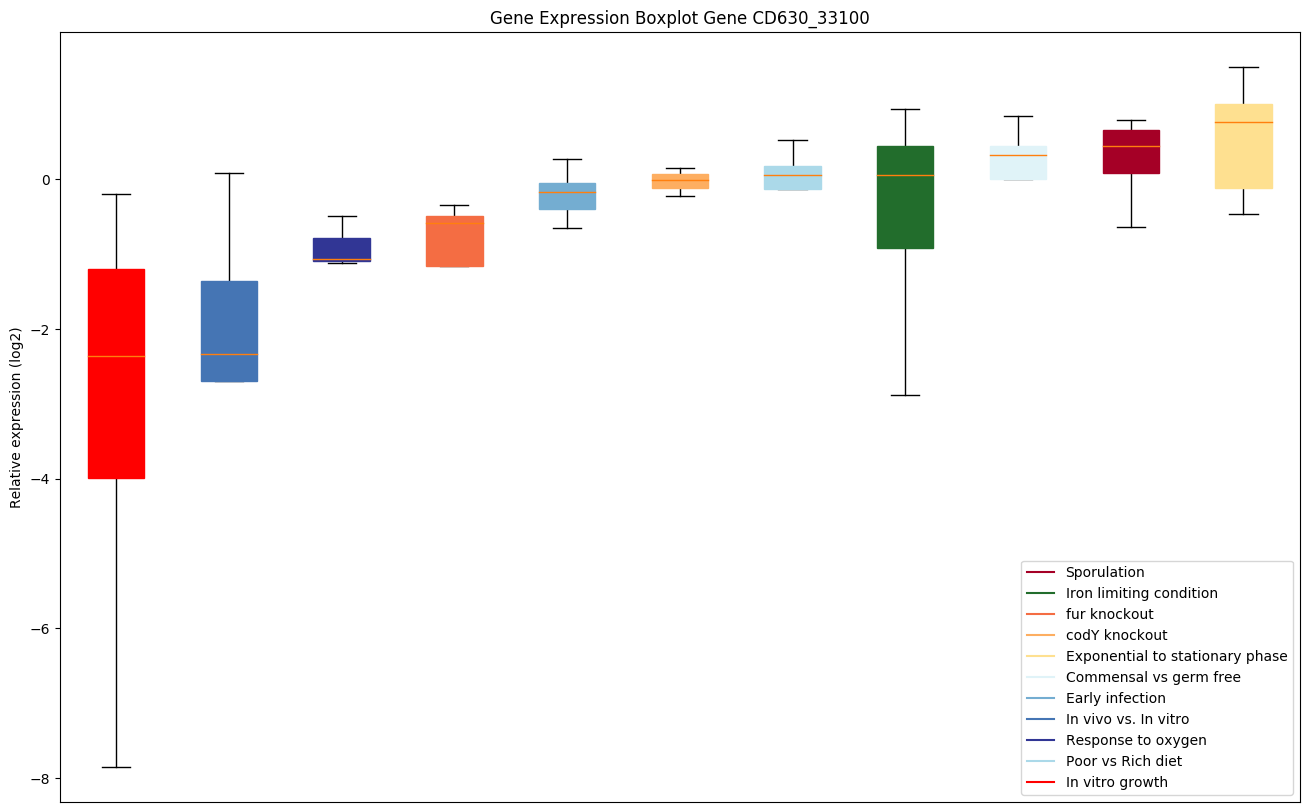
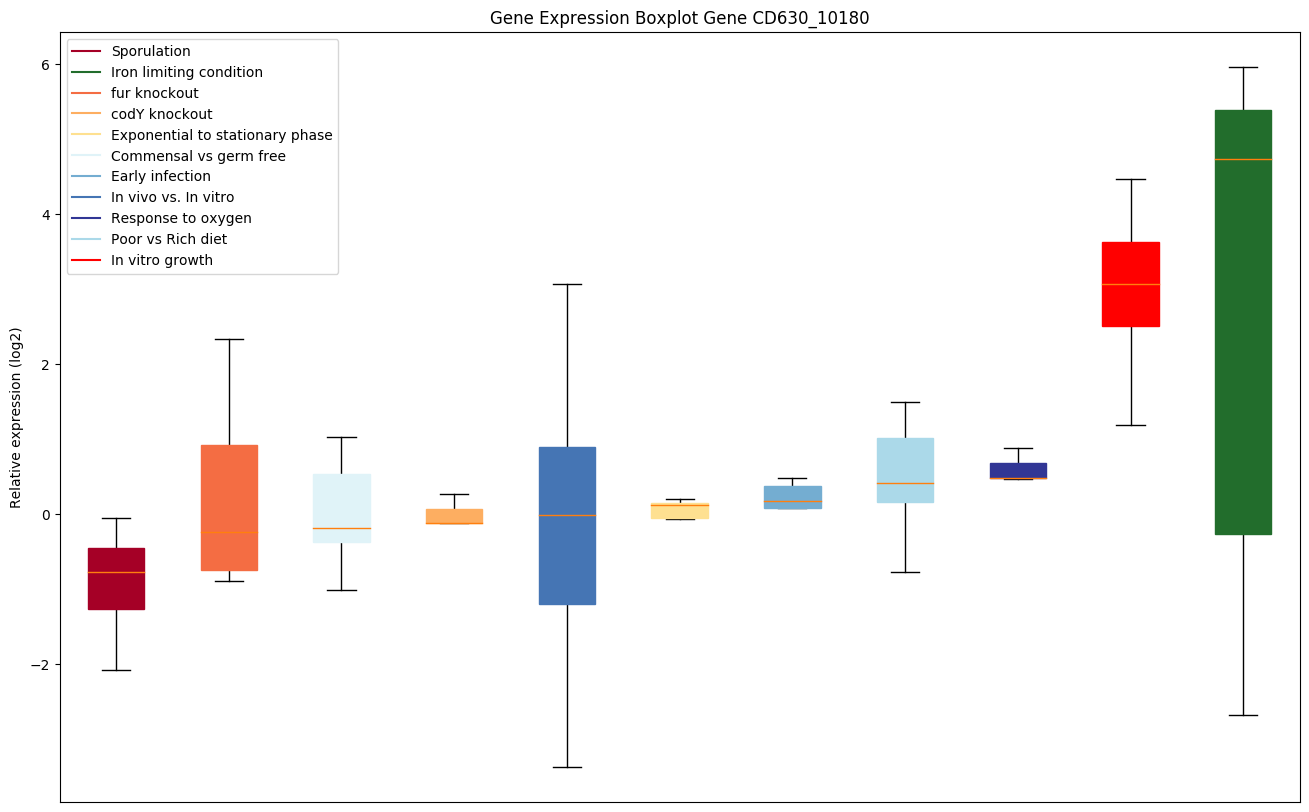
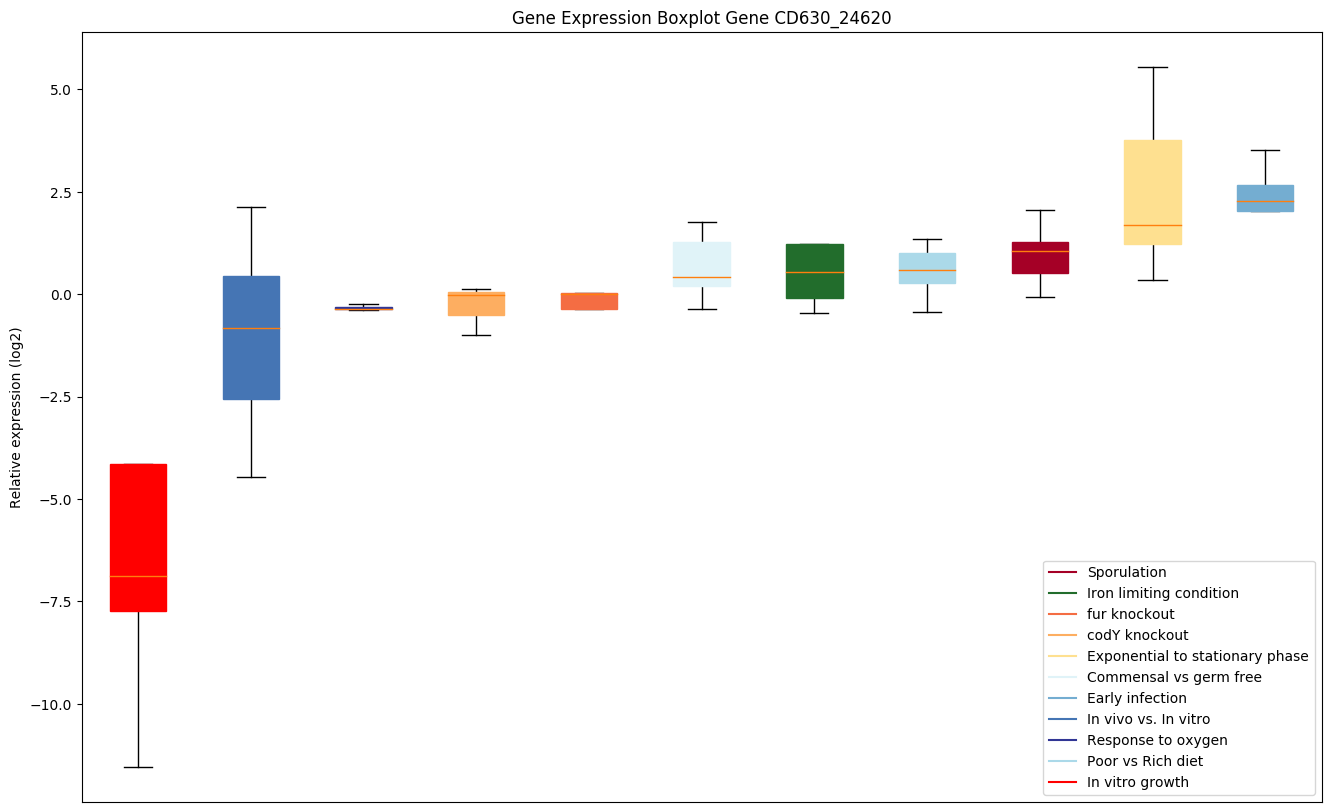
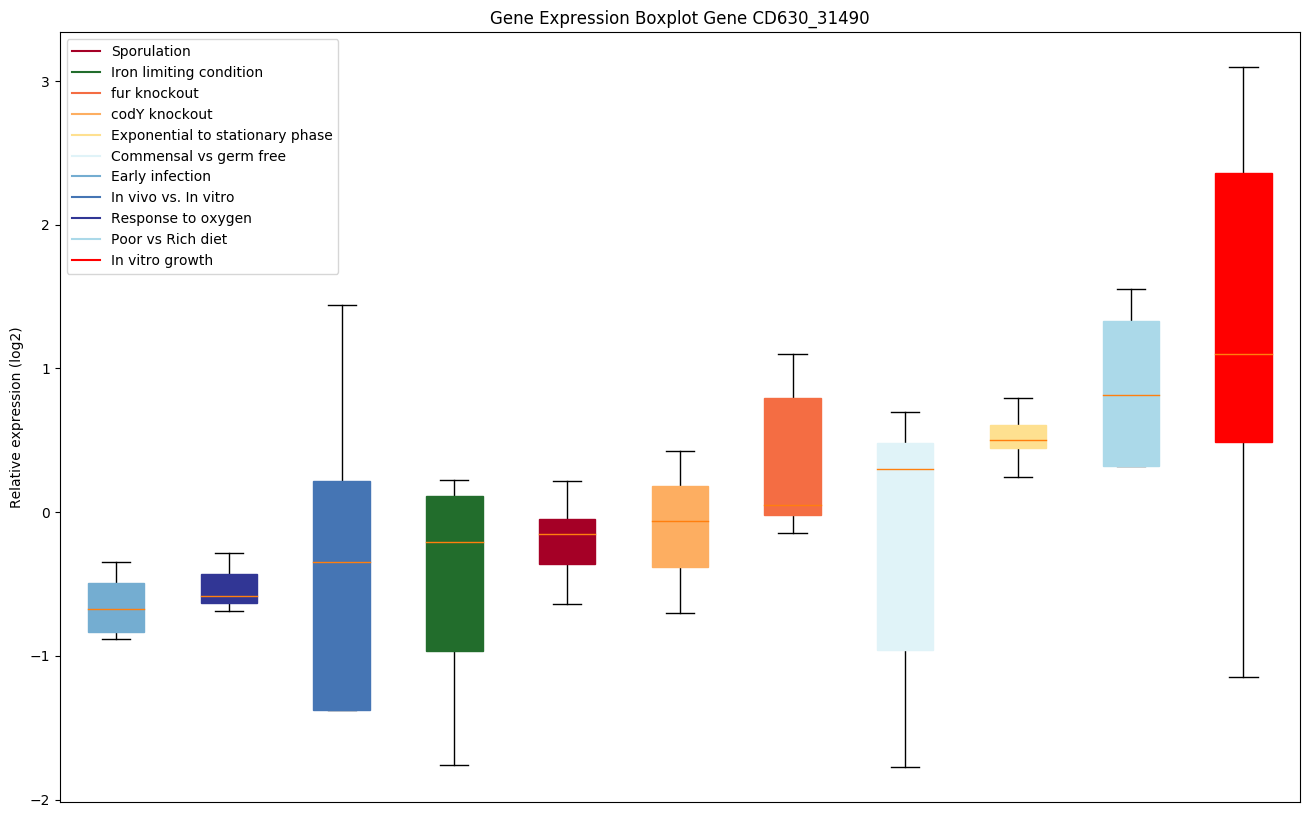
Bicluster expression profile
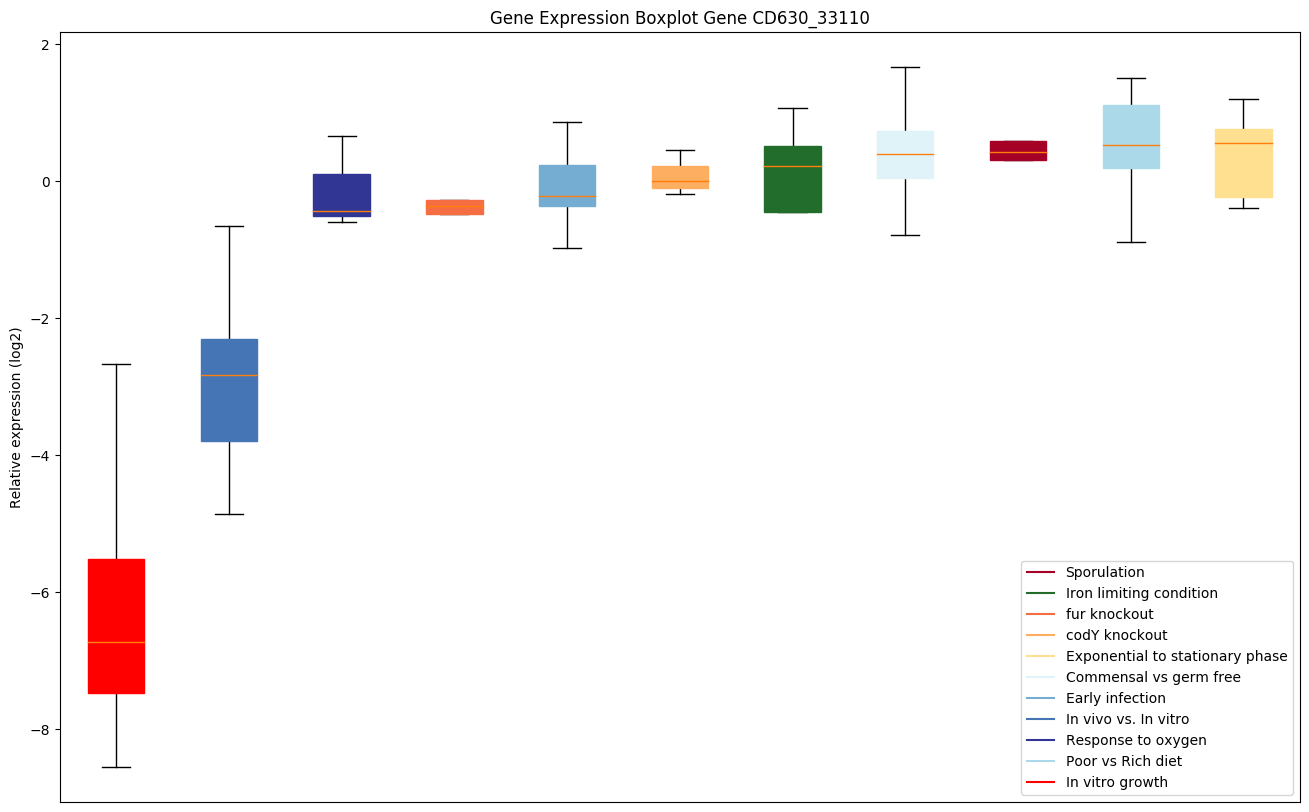
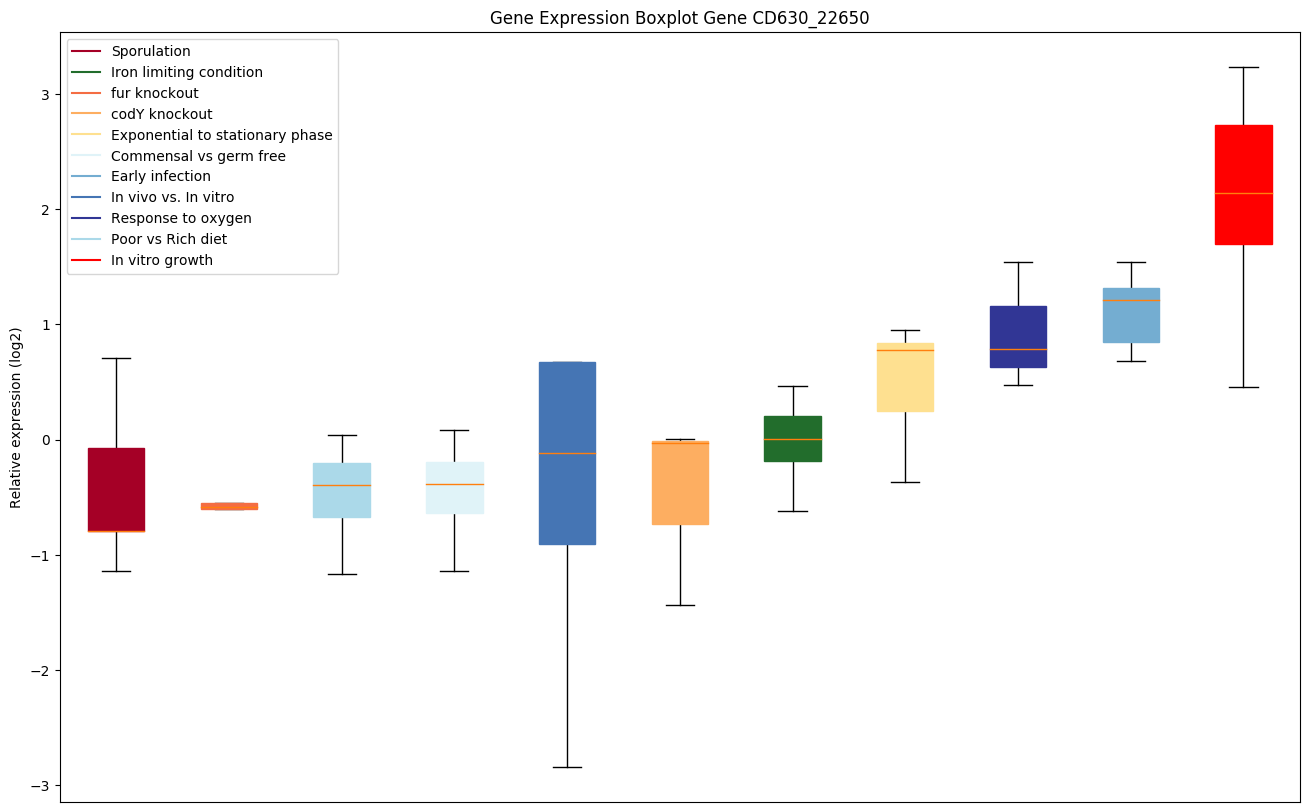
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
The module is significantly enriched with genes associated to the indicated functional terms
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_20350 | Putative sporulation integral membrane protein |
 |
|||||
| CD630_18591 | Transcriptional regulator, AbrB family;Tn1549-like, CTn5-Orf17 |
 |
|||||
| CD630_18590 | DNA topoisomerase protein, type IA, Tn1549-like,CTn5-Orf16 |
 |
|||||
| CD630_24610 | dnaK | Chaperone protein dnaK (Heat shock protein 70)(Heat shock 70 kDa protein) (HSP70) | Acts as a chaperone. | Yes |
 |
||
| CD630_18550 | Putative conjugative transposon proteinTn1549-like, CTn5-Orf11 |
 |
|||||
| CD630_18620 | Putative DNA/RNA helicase Tn1549-like,CTn5-Orf21 |
 |
|||||
| CD630_31480 | Hypothetical protein |
 |
|||||
| CD630_20390 | Conserved hypothetical protein |
 |
|||||
| CD630_18571 | Putative conjugative transposon proteinTn1549-like, CTn5-Orf14 |
 |
|||||
| CD630_18570 | Putative cell wall hydrolase Tn1549-like,CTn5-Orf13 |
 |
|||||
| CD630_18530 | Putative membrane protein Tn1549-like, CTn5-Orf9 |
 |
|||||
| CD630_18510 | Putative single-strand DNA-binding proteinTn1549-like, CTn5-Orf7 |
 |
|||||
| CD630_10170 | ABC-type transport system, multidrug-familyATP-binding protein/permease |
 |
|||||
| CD630_22660 | Putative oxidoreductase, FAD dependent |
 |
|||||
| CD630_33100 | Putative D-isomer specific 2-hydroxyaciddehydrogenase |
 |
|||||
| CD630_22620 | Transporter, Major Facilitator Superfamily (MFS) |
 |
|||||
| CD630_10180 | ABC-type transport system, multidrug-familyATP-binding protein/permease |
 |
|||||
| CD630_18600 | Putative conjugative transposon protein DUF1016family, Tn1549-like, CTn5-Orf18 |
 |
|||||
| CD630_18580 | Putative cell surface protein Tn1549-like,CTn5-Orf15 |
 |
|||||
| CD630_24600 | dnaJ | Chaperone protein DnaJ | Participates actively in the response to hyperosmotic and heat shock by preventing the aggregation of stress-denatured proteins and by disaggregating proteins, also in an autonomous, DnaK-independent fashion. Unfolded proteins bind initially to DnaJ; upon interaction with the DnaJ-bound protein, DnaK hydrolyzes its bound ATP, resulting in the formation of a stable complex. GrpE releases ADP from DnaK; ATP binding to DnaK triggers the release of the substrate protein, thus completing the reaction cycle. Several rounds of ATP-dependent interactions between DnaJ, DnaK and GrpE are required for fully efficient folding. Also involved, together with DnaK and GrpE, in the DNA replication of plasmids through activation of initiation proteins. |
 |
|||
| CD630_33120 | Transporter, Major Facilitator Superfamily (MFS) |
 |
|||||
| CD630_20400 | Transcriptional regulator, MerR family |
 |
|||||
| CD630_24640 | hemN | Oxygen-independent coproporphyrinogen-III oxidase(Coproporphyrinogenase) (Coprogen oxidase) | Probably acts as a heme chaperone, transferring heme to an unknown acceptor. Binds one molecule of heme per monomer, possibly covalently. Binds 1 [4Fe-4S] cluster. The cluster is coordinated with 3 cysteines and an exchangeable S-adenosyl-L-methionine. |
 |
|||
| CD630_24620 | grpE | Protein grpE (HSP-70 cofactor) | Participates actively in the response to hyperosmotic and heat shock by preventing the aggregation of stress-denatured proteins, in association with DnaK and GrpE. It is the nucleotide exchange factor for DnaK and may function as a thermosensor. Unfolded proteins bind initially to DnaJ; upon interaction with the DnaJ-bound protein, DnaK hydrolyzes its bound ATP, resulting in the formation of a stable complex. GrpE releases ADP from DnaK; ATP binding to DnaK triggers the release of the substrate protein, thus completing the reaction cycle. Several rounds of ATP-dependent interactions between DnaJ, DnaK and GrpE are required for fully efficient folding. | Yes |
 |
||
| CD630_20360 | Putative peptidylarginine deiminase |
 |
|||||
| CD630_31490 | DNA helicase |
 |
|||||
| CD630_18540 | Putative conjugative transposon proteinTn1549-like, CTn5-Orf10 |
 |
|||||
| CD630_19941 | Transcriptional regulator, ArsR family |
 |
|||||
| CD630_19940 | Putative membrane protein, DUF1648 family |
 |
|||||
| CD630_18560 | Putative hydrolase Tn1549-like, CTn5-Orf12 |
 |
|||||
| CD630_18610 | Putative conjugative transposon proteinTn1549-like, CTn5-Orf19 |
 |
|||||
| CD630_20380 | Putative drug/sodium antiporter, MATE family |
 |
|||||
| CD630_18520 | Putative conjugative transposon proteinTn1549-like, CTn5-Orf8 |
 |
|||||
| CD630_36140 | Conserved hypothetical protein, DUF1130 family |
 |
|||||
| CD630_05650 | nth | Endonuclease III | DNA repair enzyme that has both DNA N-glycosylase activity and AP-lyase activity. The DNA N-glycosylase activity releases various damaged pyrimidines from DNA by cleaving the N-glycosidic bond, leaving an AP (apurinic/apyrimidinic) site. The AP-lyase activity cleaves the phosphodiester bond 3' to the AP site by a beta-elimination, leaving a 3'-terminal unsaturated sugar and a product with a terminal 5'-phosphate. |
 |
|||
| CD630_33110 | Conserved hypothetical protein |
 |
|||||
| CD630_22650 | Putative membrane protein |
 |
|||||
| CD630_18630 | Putative single-strand DNA-binding proteinTn1549-like, CTn5-Orf22 |
 |
|||||
| CD630_20370 | Putative acyl-CoA acyltransferase |
 |
|||||
| CD630_31500 | Conserved hypothetical protein |
 |