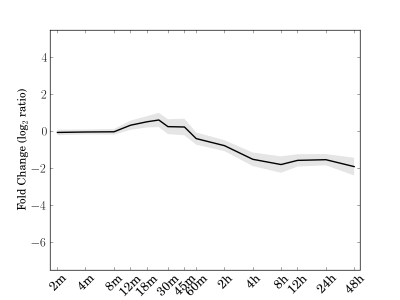
Module 99 Residual: 0.15
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0099 | v02 | 0.15 | -12.89 |
Displaying 1 - 19 of 19
| Cre01.g033400.t1.2 Tim10/DDP family zinc finger protein | Cre01.g037700.t1.2 Fatty acid/sphingolipid desaturase |
| Cre02.g080700.t1.2 Heat shock protein 70 (Hsp 70) family protein | Cre02.g084350.t1.2 Uncharacterized protein family (UPF0016) |
| Cre02.g097400.t1.2 eukaryotic elongation factor 5A-1 | Cre03.g188700.t1.2 Plastid-lipid associated protein PAP / fibrillin family protein |
| Cre05.g230850.t1.1 | Cre05.g245102.t1.1 |
| Cre06.g250100.t1.2 chloroplast heat shock protein 70-2 | Cre06.g253350.t1.2 Single hybrid motif superfamily protein |
| Cre06.g267600.t2.1 Lycopene beta/epsilon cyclase protein | Cre07.g339700.t1.2 non-intrinsic ABC protein 7 |
| Cre07.g340200.t1.1 PGR5-like B | Cre08.g372950.t1.2 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
| Cre12.g500750.t1.2 | Cre12.g537641.t1.1 MBOAT (membrane bound O-acyl transferase) family protein |
| Cre13.g604300.t1.2 | Cre16.g684300.t1.2 YGGT family protein |
| Cre17.g726450.t1.2 |
|
e.value: 0.026 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: tGGtGgTGGCgGnGGCgGtGG |
motif_0099_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 14 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: atacTaTacTTTtTT |
Displaying 1 - 5 of 5
| Interaction | Weight | |
|---|---|---|
| down-regulates |
Cre03.g149350.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51RWP-RK domain-containing protein |
0.046053 |
| down-regulates |
Cre10.g453500.t1.2Submitted by admin on Tue, 05/19/2015 - 15:51 |
0.059203 |
| down-regulates |
Cre12.g523000.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51zinc finger protein 1 |
0.044403 |
| down-regulates |
Cre08.g375400.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51KNOTTED-like homeobox of Arabidopsis thaliana 7 |
0.060887 |
| up-regulates |
Cre12.g534450.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51SMAD/FHA domain-containing protein |
0.064694 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0006629 | lipid metabolic process |

Comments