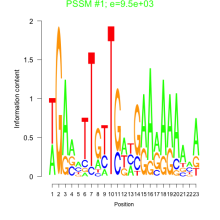
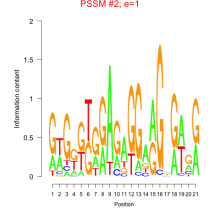
Thaps_bicluster_0020 Residual: 0.47
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0020 | 0.47 | Thalassiosira pseudonana |
Displaying 1 - 56 of 56
" class="views-fluidgrid-wrapper clear-block">
1135 hypothetical protein
11391 hypothetical protein
11612 AFD_class_I superfamily
12130 hypothetical protein
1444 hypothetical protein
18775 MORN superfamily
19094 S_TKc
19622 Sedlin_N
19637 hypothetical protein
20630 Piwi-like superfamily
2066 hypothetical protein
20992 hypothetical protein
24425 RhaT
24576 hypothetical protein
25159 hypothetical protein
25551 hypothetical protein
25702 hypothetical protein
261602 ThiF_MoeB_HesA_family
2671 Nudix_Hydrolase_31
269666 MPN superfamily
269690 (Tp_Hox1) regulator [Rayko]
269705 hypothetical protein
269844 GT1_Gtf_like
270240 hypothetical protein
3035 hypothetical protein
32561 (Tp_CCHH4) DnaJ
33067 (Tp_CBF/NF4) CBFD_NFYB_HMF
34344 Mito_carr
35182 SET
3708 hypothetical protein
37707 hypothetical protein
38194 MPP_PhoA_N
39143 (ANT1) PTZ00169
40391 (ENO1) PLN00191
406 (GOX) FMN_dh
4084 hypothetical protein
42965 GlcD
4971 hypothetical protein
5579 Hydrolase_4 superfamily
6657 hypothetical protein
6768 DUF726 superfamily
6856 (Tp_HSF_1.3f) HSF_DNA-bind superfamily
7202 ABCC_MRP_domain1
7480 hypothetical protein
8092 hypothetical protein
8636 hypothetical protein
875 Trypsin_2
ThpsCp002 (psaB) photosystem I P700 chlorophyll a apoprotein A2
ThpsCp026 (ycf45) hypothetical protein
ThpsCp027 (dnaB) DNA replication helicase
ThpsCp052 (psaI) photosystem I subunit VIII
ThpsCp077 (rpl32) ribosomal protein L32
ThpsCp078 (rbcR) LysR transcriptional regulator
ThpsCp125 (clpC) Clp protease ATP binding subunit
ThpsCp138 (rbcR) LysR transcriptional regulator
ThpsCt028 (trnL(uag)')
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009166 | nucleotide catabolism | 0.00701752 | 1 | Thaps_bicluster_0020 |
| GO:0006888 | ER to Golgi transport | 0.0209137 | 1 | Thaps_bicluster_0020 |
| GO:0006725 | aromatic compound metabolism | 0.0746894 | 1 | Thaps_bicluster_0020 |
| GO:0006096 | glycolysis | 0.122569 | 1 | Thaps_bicluster_0020 |
| GO:0006810 | transport | 0.134855 | 1 | Thaps_bicluster_0020 |
| GO:0000074 | regulation of cell cycle | 0.191478 | 1 | Thaps_bicluster_0020 |
| GO:0006118 | electron transport | 0.23943 | 1 | Thaps_bicluster_0020 |
| GO:0006508 | proteolysis and peptidolysis | 0.268722 | 1 | Thaps_bicluster_0020 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.294119 | 1 | Thaps_bicluster_0020 |
| GO:0006457 | protein folding | 0.371189 | 1 | Thaps_bicluster_0020 |

Comments