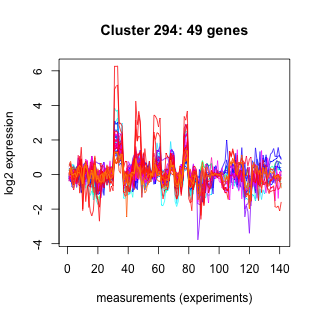
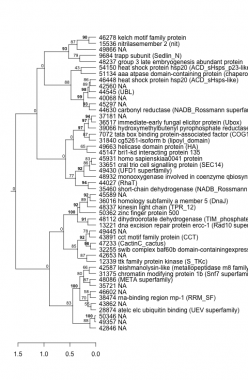
Phatr_hclust_0294 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006207 | 'de novo' pyrimidine base biosynthesis | 0.0225371 | 1 | Phatr_hclust_0294 |
| GO:0006888 | ER to Golgi transport | 0.0445878 | 1 | Phatr_hclust_0294 |
| GO:0006464 | protein modification | 0.0488856 | 1 | Phatr_hclust_0294 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 0.0608128 | 1 | Phatr_hclust_0294 |
| GO:0007155 | cell adhesion | 0.0714826 | 1 | Phatr_hclust_0294 |
| GO:0006807 | nitrogen compound metabolism | 0.076774 | 1 | Phatr_hclust_0294 |
| GO:0008152 | metabolism | 0.0807231 | 1 | Phatr_hclust_0294 |
| GO:0006725 | aromatic compound metabolism | 0.107921 | 1 | Phatr_hclust_0294 |
| GO:0015031 | protein transport | 0.162433 | 1 | Phatr_hclust_0294 |
| GO:0006281 | DNA repair | 0.176738 | 1 | Phatr_hclust_0294 |
|
PHATRDRAFT_32255 : swib complex baf60b domain-containingexpressed (SWIB) |
PHATRDRAFT_48932 : monooxygenase involved in coenzyme qbiosynthesis (NAD_binding_8 superfamily) |
||
|
PHATRDRAFT_47233 : (CactinC_cactus) |
PHATRDRAFT_49430 : (UFD1 superfamily) |
||
|
PHATRDRAFT_43891 : cct motif family protein (CCT) |
PHATRDRAFT_33651 : cral trio cell signalling protein (SEC14) |
PHATRDRAFT_44545 : (UBL) |
|
|
PHATRDRAFT_28874 : atelc elc ubiquitin binding (UEV superfamily) |
PHATRDRAFT_45931 : homo sapienskiaa0041 protein |
||
|
PHATRDRAFT_13221 : dna excision repair protein ercc-1 (Rad10 superfamily) |
PHATRDRAFT_45147 : bri1-kd interacting protein 135 |
PHATRDRAFT_46448 : heat shock protein hsp20 (ACD_sHsps-like) |
|
|
PHATRDRAFT_38474 : rna-binding region rnp-1 (RRM_SF) |
PHATRDRAFT_48112 : dihydroorotate dehydrogenase (TIM_phosphate_binding superfamily) |
PHATRDRAFT_49663 : helicase domain protein (HA) |
PHATRDRAFT_51134 : aaa atpase domain-containing protein (chaperone_ClpB) |
|
PHATRDRAFT_50362 : zinc finger protein 500 |
PHATRDRAFT_31840 : cg5261-isoform b (lipoyl_domain) |
PHATRDRAFT_54150 : heat shock protein hsp20 (ACD_sHsps_p23-like) |
|
|
PHATRDRAFT_48337 : kinesin light chain (TPR_12) |
PHATRDRAFT_7072 : tata box binding protein-associated factor (COG1936) |
PHATRDRAFT_48237 : group 3 late embryogenesis abundant protein |
|
|
PHATRDRAFT_48086 : (META superfamily) |
PHATRDRAFT_36016 : homology subfamily a member 5 (DnaJ) |
PHATRDRAFT_39066 : hydroxymethylbutenyl pyrophosphate reductase (S1_like superfamily) |
PHATRDRAFT_9684 : trapp subunit (Sedlin_N) |
|
PHATRDRAFT_31375 : chromatin modifying protein 1b (Snf7 superfamily) |
PHATRDRAFT_36517 : immediate-early fungal elicitor protein (Ubox) |
||
|
PHATRDRAFT_42587 : leishmanolysin-like (metallopeptidase m8 family) (Peptidase_M8 superfamily) |
PHATRDRAFT_35460 : short-chain dehydrogenase (NADB_Rossmann superfamily) |
PHATRDRAFT_15536 : nitrilasemember 2 (nit) |
|
|
PHATRDRAFT_12339 : ttk family protein kinase (S_TKc) |
PHATRDRAFT_44027 : (RhaT) |
PHATRDRAFT_44630 : carbonyl reductase (NADB_Rossmann superfamily) |
PHATRDRAFT_46278 : kelch motif family protein |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | -0.261 | 0.0674208 |
| Copper_SH | Copper_SH | -0.211 | 0.00431655 |
| Green_vs_Red_24h | Green_vs_Red_24h | -0.029 | 0.795725 |
| Silver_SH | Silver_SH | -0.079 | 0.262647 |
| Cadmium_1.2mg | Cadmium_1.2mg | 0.008 | 0.925278 |
| BlueLight_24h | BlueLight_24h | -0.156 | 0.190994 |
| GreenLight_0.5h | GreenLight_0.5h | 0.679 | 0.00310077 |
| RedLight_6h | RedLight_6h | -0.348 | 0.0180769 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 1 |
| GreenLight_24h | GreenLight_24h | -0.145 | 0.212363 |
| light_6hr | light_6hr | 0.453 | 0.00192308 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | 0.366 | 0.005 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | -0.056 | 0.446347 |
| Ammonia_SH | Ammonia_SH | -0.696 | 0.000662252 |
| Simazine_SH | Simazine_SH | 0.022 | 0.801789 |
| Oil_SH | Oil_SH | -0.162 | 0.0336898 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | 0.041 | 0.616552 |
| Cadmium_SH | Cadmium_SH | -0.198 | 0.0200549 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | -0.041 | 0.772207 |
| highlight_0to6h | highlight_0to6h | 0.051 | 0.823028 |
| BlueLight_0.5h | BlueLight_0.5h | 1.353 | 0.000462963 |
| Dispersant_SH | Dispersant_SH | 0.236 | 0.0056962 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | 0.019 | 0.848894 |
| light_15.5hr | light_15.5hr | 0.155 | 0.377855 |
| light_10.5hr | light_10.5hr | 0.596 | 0.000561798 |
| Dispersed_oil_SH | Dispersed_oil_SH | -0.008 | 0.926235 |
| Si_free | Si_free | -0.026 | 0.776005 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | 1.396 | 0.000396825 |
| Mixture_SH | Mixture_SH | -0.778 | 0.000357143 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | -0.081 | 0.641549 |
| light_16hr_dark_30min | light_16hr_dark_30min | -0.003 | 0.992581 |
| BlueLight_6h | BlueLight_6h | -0.429 | 0.000581395 |
| highlight_12to48h | highlight_12to48h | -0.241 | 0.0685096 |
| RedLight_24h | RedLight_24h | -0.116 | 0.324721 |
| Dark_treated | Dark_treated | 1.940 | 0.000446429 |
| GreenLight_6h | GreenLight_6h | -0.372 | 0.00390625 |
| Cadmium_0.12mg | Cadmium_0.12mg | -0.019 | 0.544792 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | -0.012 | 0.887799 |
| RedLight_0.5h | RedLight_0.5h | 0.646 | 0.00795455 |
| Green_vs_Red_6h | Green_vs_Red_6h | -0.025 | 0.894226 |
| Re-illuminated_24h | Re-illuminated_24h | -0.165 | 0.0816667 |