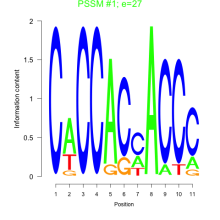
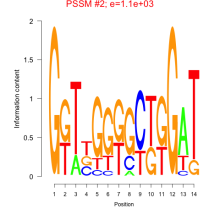
Thaps_bicluster_0136 Residual: 0.28
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0136 | 0.28 | Thalassiosira pseudonana |
Displaying 1 - 21 of 21
" class="views-fluidgrid-wrapper clear-block">
10353 hypothetical protein
1038 SPRYD7
13117 GTPBP1
1850 PKc_like superfamily
2086 zf-UBP
2319 hypothetical protein
263219 MFS_1
263399 CypX
263558 Menin superfamily
263810 PTZ00430
264157 CBS_pair superfamily
2702 Pro_isomerase
28148 (NIT2) nit
35956 SIR2 superfamily
3837 UshA
3913 LCM
4456 Abhydrolase_6
5038 DUF179
5335 Cyt_b561 superfamily
6140 hypothetical protein
7516 Aldo_ket_red
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009166 | nucleotide catabolism | 0.00454357 | 1 | Thaps_bicluster_0136 |
| GO:0006342 | chromatin silencing | 0.0202987 | 1 | Thaps_bicluster_0136 |
| GO:0006094 | gluconeogenesis | 0.0202987 | 1 | Thaps_bicluster_0136 |
| GO:0006807 | nitrogen compound metabolism | 0.0247583 | 1 | Thaps_bicluster_0136 |
| GO:0006334 | nucleosome assembly | 0.0683505 | 1 | Thaps_bicluster_0136 |
| GO:0006096 | glycolysis | 0.0810783 | 1 | Thaps_bicluster_0136 |
| GO:0006457 | protein folding | 0.25918 | 1 | Thaps_bicluster_0136 |
| GO:0006412 | protein biosynthesis | 0.319237 | 1 | Thaps_bicluster_0136 |
| GO:0006468 | protein amino acid phosphorylation | 0.39098 | 1 | Thaps_bicluster_0136 |
| GO:0006118 | electron transport | 0.463588 | 1 | Thaps_bicluster_0136 |

Comments