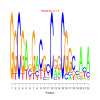
Module 24 Residual: 0.54
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0024 | v02 | 0.54 | -17.77 |
| Rv1744c | Rv2701c Inositol-1-monophosphatase (EC 3.1.3.25) |
| Rv2702 Polyphosphate glucokinase (EC 2.7.1.63) | Rv2748c Cell division protein FtsK |
| Rv3331 Sugar-transport integral membrane proteinSugI | Rv3332 N-acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25) |
| Rv3439c Conserved alanine and proline rich protein | Rv3440c |
| Rv3904c Esat-6 like protein EsxE | Rv2311 Putative ATP-dependent exoDNAse |
|
GO:0000287 | magnesium ion binding |
|
GO:0008934 | inositol monophosphate 1-phosphatase activity |
|
GO:0051260 | protein homooligomerization |
|
GO:0047330 | polyphosphate-glucose phosphotransferase activity |
|
GO:0004340 | glucokinase activity |
|
GO:0040007 | growth |
|
GO:0008150 | biological_process |
|
GO:0005576 | extracellular region |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0005365 | myo-inositol transmembrane transporter activity |
|
GO:0008448 | N-acetylglucosamine-6-phosphate deacetylase activity |












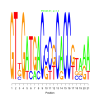
Discussion