Module 65 Residual: 0.72
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0065 | v02 | 0.72 | -10.60 |
| Rv0315 \Possible beta-1,3-glucanase\\\"" | Rv0760c steroid isomerase, putative |
| Rv0761c Alcohol dehydrogenase (EC 1.1.1.1) | Rv1078 Proline-rich antigen homolog |
| Rv1241 | Rv1242 |
| Rv1589 Biotin synthase (EC 2.8.1.6) | Rv1590 |
| Rv1591 Possible membrane protein | Rv2213 Cytosol aminopeptidase PepA (EC 3.4.11.1) |
| Rv2911 D-alanyl-D-alanine carboxypeptidase (EC 3.4.16.4) | Rv2929 |
| Rv3791 3-oxoacyl-[acyl-carrier protein] reductase paralog (EC 1.1.1.100) | Rv3794 Probable arabinosyltransferase A (EC 2.4.2.-) |
| Rv3795 INTEGRAL MEMBRANE INDOLYLACETYLINOSITOL ARABINOSYLTRANSFERASE EMBC (ARABINOSYLINDOLYLACETYLINOSITOL SYNTHASE) | Rv2211c Aminomethyltransferase (glycine cleavage system T protein) (EC 2.1.2.10) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0008194 | UDP-glycosyltransferase activity |
|
GO:0005576 | extracellular region |
|
GO:0042803 | protein homodimerization activity |
|
GO:0005886 | plasma membrane |
|
GO:0045927 | positive regulation of growth |
|
GO:0045926 | negative regulation of growth |
|
GO:0004076 | biotin synthase activity |
|
GO:0005887 | integral to plasma membrane |
|
GO:0004178 | leucyl aminopeptidase activity |
|
GO:0009002 | serine-type D-Ala-D-Ala carboxypeptidase activity |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0040007 | growth |
|
GO:0005829 | cytosol |
|
GO:0016763 | transferase activity, transferring pentosyl groups |
|
GO:0005618 | cell wall |


















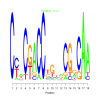
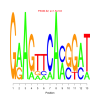
Discussion