Module 129 Residual: 0.53
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0129 | v02 | 0.53 | -19.81 |
| Rv1221 RNA polymerase sigma-70 factor, ECF subfamily | Rv1536 Isoleucyl-tRNA synthetase (EC 6.1.1.5) |
| Rv2009 | Rv2697c Deoxyuridine 5'-triphosphate nucleotidohydrolase (EC 3.6.1.23) |
| Rv2698 PROBABLE CONSERVED ALANINE RICH TRANSMEMBRANE PROTEIN | Rv3106 Ferredoxin--NADP(+) reductase, actinobacterial (eukaryote-like) type (EC 1.18.1.2) |
| Rv3854c monooxygenase, flavin-binding family | Rv3855 Transcriptional repressor EthR, TetR family |
| Rv3105c Peptide chain release factor 2 |
|
GO:0003677 | DNA binding |
|
GO:0009405 | pathogenesis |
|
GO:0009408 | response to heat |
|
GO:0009410 | response to xenobiotic stimulus |
|
GO:0016987 | sigma factor activity |
|
GO:0042542 | response to hydrogen peroxide |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0052572 | response to host immune response |
|
GO:0004822 | isoleucine-tRNA ligase activity |
|
GO:0005618 | cell wall |
|
GO:0005886 | plasma membrane |
|
GO:0001666 | response to hypoxia |
|
GO:0045927 | positive regulation of growth |
|
GO:0004170 | dUTP diphosphatase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0040007 | growth |
|
GO:0046080 | dUTP metabolic process |
|
GO:0004324 | ferredoxin-NADP+ reductase activity |
|
GO:0008860 | ferredoxin-NAD+ reductase activity |
|
GO:0050660 | flavin adenine dinucleotide binding |
|
GO:0055114 | oxidation-reduction process |
|
GO:0070401 | NADP+ binding |
|
GO:0004499 | N,N-dimethylaniline monooxygenase activity |
|
GO:0005829 | cytosol |
|
GO:0042803 | protein homodimerization activity |
|
GO:0045892 | negative regulation of transcription, DNA-dependent |
|
GO:0046677 | response to antibiotic |
|
GO:0051260 | protein homooligomerization |
|
GO:0006415 | translational termination |
|
GO:0003747 | translation release factor activity |











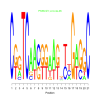
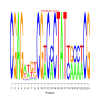
Discussion