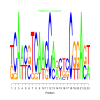
Module 308 Residual: 0.49
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0308 | v02 | 0.49 | -23.44 |
| Rv1338 Glutamate racemase (EC 5.1.1.3) | Rv2684 Na+/H+ antiporter NhaD type |
| Rv2685 Na+/H+ antiporter NhaD type | Rv3719 FAD/FMN-containing dehydrogenases |
| Rv3720 Cyclopropane-fatty-acyl-phospholipid synthase (EC 2.1.1.79) | Rv1334 Mec+ |
| Rv1336 Cysteine synthase B (EC 2.5.1.47) | Rv1337 Rhomboid membrane family protein |
| Rv1769 L-gulono-1,4-lactone oxidase (EC 1.1.3.8) | Rv1770 Putative aminopeptidase |
| Rv1771 oxidoreductase, FAD-binding | Rv2148c |
| Rv2149c COG1496: Uncharacterized conserved protein | Rv2682c 1-deoxy-D-xylulose 5-phosphate synthase (EC 2.2.1.7) |
| Rv2683 | Rv3733c MutT-like protein |
| Rv3734c Wax ester synthase/acyl-CoA:diacylglycerol acyltransferase | Rv3735 Probable transmembrane protein |
|
GO:0008881 | glutamate racemase activity |
|
GO:0008150 | biological_process |
|
GO:0005515 | protein binding |
|
GO:0005886 | plasma membrane |
|
GO:0008657 | DNA topoisomerase (ATP-hydrolyzing) inhibitor activity |
|
GO:0008235 | metalloexopeptidase activity |
|
GO:0008270 | zinc ion binding |
|
GO:0019344 | cysteine biosynthetic process |
|
GO:0004124 | cysteine synthase activity |
|
GO:0030170 | pyridoxal phosphate binding |
|
GO:0033847 | O-phosphoserine sulfhydrylase activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0043234 | protein complex |
|
GO:0005618 | cell wall |
|
GO:0080049 | L-gulono-1,4-lactone dehydrogenase activity |
|
GO:0008661 | 1-deoxy-D-xylulose-5-phosphate synthase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0019288 | isopentenyl diphosphate biosynthetic process, mevalonate-independent pathway |
|
GO:0030145 | manganese ion binding |
|
GO:0030975 | thiamine binding |
|
GO:0040007 | growth |
|
GO:0001666 | response to hypoxia |
|
GO:0004144 | diacylglycerol O-acyltransferase activity |
|
GO:0045017 | glycerolipid biosynthetic process |
|
GO:0071731 | response to nitric oxide |




















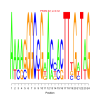
Discussion