Module 493 Residual: 0.66
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0493 | v02 | 0.66 | -13.45 |
| Rv0297 PE-PGRS family protein | Rv0126 Trehalose synthase (EC 5.4.99.16) |
| Rv0141c | Rv0156 NAD(P) transhydrogenase alpha subunit (EC 1.6.1.2) |
| Rv0163 putative 4-hydroxybenzoyl-CoA thioesterase | Rv0295c Sulfotransferase |
| Rv0296c Choline-sulfatase (EC 3.1.6.6) | Rv0302 TetR/ACRR family transcriptional regulator |
| Rv0303 \Oxidoreductase, short-chain dehydrogenase/reductase family\\\"" | Rv0609 |
| Rv0617 | Rv0812 Aminodeoxychorismate lyase (EC 4.1.3.38) |
| Rv0866 Molybdenum cofactor biosynthesis protein MoaE | Rv1009 Cell wall-binding protein |
| Rv1010 Dimethyladenosine transferase (EC 2.1.1.-) | Rv1011 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (EC 2.7.1.148) |
| Rv1897c D-tyrosyl-tRNA(Tyr) deacylase | Rv2976c Uracil-DNA glycosylase, family 1 |
| Rv3123 | Rv3634c UDP-glucose 4-epimerase (EC 5.1.3.2) |
|
GO:0005886 | plasma membrane |
|
GO:0005978 | glycogen biosynthetic process |
|
GO:0005992 | trehalose biosynthetic process |
|
GO:0016161 | beta-amylase activity |
|
GO:0047471 | maltose alpha-D-glucosyltransferase activity |
|
GO:0008750 | NAD(P)+ transhydrogenase (AB-specific) activity |
|
GO:0005618 | cell wall |
|
GO:0005991 | trehalose metabolic process |
|
GO:0008146 | sulfotransferase activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0046506 | sulfolipid biosynthetic process |
|
GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process |
|
GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding |
|
GO:0045926 | negative regulation of growth |
|
GO:0008696 | 4-amino-4-deoxychorismate lyase activity |
|
GO:0005515 | protein binding |
|
GO:0005576 | extracellular region |
|
GO:0010628 | positive regulation of gene expression |
|
GO:0010629 | negative regulation of gene expression |
|
GO:0040010 | positive regulation of growth rate |
|
GO:0008649 | rRNA methyltransferase activity |
|
GO:0000154 | rRNA modification |
|
GO:0050515 | 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase activity |
|
GO:0040007 | growth |
|
GO:0019478 | D-amino acid catabolic process |
|
GO:0005737 | cytoplasm |
|
GO:0004844 | uracil DNA N-glycosylase activity |
|
GO:0003978 | UDP-glucose 4-epimerase activity |






















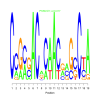

Discussion