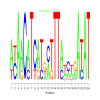
Module 541 Residual: 0.44
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0541 | v02 | 0.44 | -19.01 |
| Rv0165c Predicted regulator PutR for proline utilization, GntR family | Rv0166 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) |
| Rv0167 | Rv0169 MCE-family protein Mce1A |
| Rv0170 MCE-family protein Mce1B | Rv0171 MCE-family protein Mce1C |
| Rv0172 MCE-family protein Mce1D | Rv0173 MCE-family lipoprotein LprK (MCE-family lipoprotein Mce1e) |
| Rv0174 MCE-family protein Mce1F | Rv0175 FIG033430: Probable conserved MCE associated membrane protein |
| Rv0177 FIG034772: Probable conserved MCE associated protein | Rv0178 FIG030769: Probable conserved MCE associated membrane protein |
| Rv0655 Methionine ABC transporter ATP-binding protein | Rv1697 FIG005773: conserved membrane protein ML1361 |
| Rv1698 | Rv3145 NADH ubiquinone oxidoreductase chain A (EC 1.6.5.3) |
| Rv3158 NADH-ubiquinone oxidoreductase chain N (EC 1.6.5.3) |
|
GO:0075182 | negative regulation of symbiont transcription in response to host |
|
GO:0005886 | plasma membrane |
|
GO:0003779 | actin binding |
|
GO:0005618 | cell wall |
|
GO:0007157 | heterophilic cell-cell adhesion |
|
GO:0008017 | microtubule binding |
|
GO:0030260 | entry into host cell |
|
GO:0042785 | active evasion of host immune response via regulation of host cytokine network |
|
GO:0044117 | growth of symbiont in host |
|
GO:0044406 | adhesion to host |
|
GO:0044409 | entry into host |
|
GO:0005576 | extracellular region |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0052572 | response to host immune response |
|
GO:0005887 | integral to plasma membrane |
|
GO:0040007 | growth |
|
GO:0006811 | ion transport |
|
GO:0009279 | cell outer membrane |
|
GO:0015288 | porin activity |
|
GO:0019867 | outer membrane |
|
GO:0046930 | pore complex |
|
GO:0008137 | NADH dehydrogenase (ubiquinone) activity |



















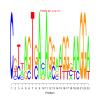
Discussion