Module 593 Residual: 0.58
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0593 | v02 | 0.58 | -7.13 |
| Rv0245 Nitrilotriacetate monooxygenase component B (EC 1.14.13.-) | Rv0474 \Transcriptional regulator, XRE family\\\"" |
| Rv0475 Iron-regulated heparin binding hemagglutinin HbhA (Adhesin) | Rv0711 Arylsulfatase (EC 3.1.6.1) |
| Rv0805 3',5'-cyclic-nucleotide phosphodiesterase (EC 3.1.4.17) | Rv0983 Heat shock protein HtrA |
| Rv0984 Molybdenum cofactor biosynthesis protein MoaB | Rv1245c Short-chain type dehydrogenase/reductase |
| Rv1531 Carboxymuconolactone decarboxylase | Rv1732c Alkyl hydroperoxide reductase and/or thiol-specific antioxidant family (AhpC/TSA) protein |
| Rv2373c Chaperone protein DnaJ | Rv2728c |
| Rv2729c Putative conserved alanine, valine and leucine rich integral membrane protein | Rv3009c Aspartyl-tRNA(Asn) amidotransferase subunit B (EC 6.3.5.6) @ Glutamyl-tRNA(Gln) amidotransferase subunit B (EC 6.3.5.7) |
| Rv3138 COG1180: Radical SAM, Pyruvate-formate lyase-activating enzyme like | Rv3139 Acyl-CoA dehydrogenase, short-chain specific (EC 1.3.99.2) |
| Rv3140 Probable acyl-CoA dehydrogenase (EC 1.3.99.3) | Rv3645 Adenylate cyclase (EC 4.6.1.1) |
| Rv3879c RD1 region associated protein Rv3879c |
|
GO:0005829 | cytosol |
|
GO:0005515 | protein binding |
|
GO:0005618 | cell wall |
|
GO:0005886 | plasma membrane |
|
GO:0007157 | heterophilic cell-cell adhesion |
|
GO:0008201 | heparin binding |
|
GO:0009986 | cell surface |
|
GO:0042803 | protein homodimerization activity |
|
GO:0044078 | positive regulation by symbiont of host receptor-mediated endocytosis |
|
GO:0010033 | response to organic substance |
|
GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity |
|
GO:0005506 | iron ion binding |
|
GO:0008199 | ferric iron binding |
|
GO:0009405 | pathogenesis |
|
GO:0030145 | manganese ion binding |
|
GO:0042301 | phosphate ion binding |
|
GO:0005576 | extracellular region |
|
GO:0031072 | heat shock protein binding |
|
GO:0006457 | protein folding |
|
GO:0051082 | unfolded protein binding |
|
GO:0040007 | growth |
|
GO:0010468 | regulation of gene expression |
|
GO:0003995 | acyl-CoA dehydrogenase activity |
|
GO:0004016 | adenylate cyclase activity |






















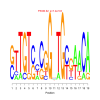
Discussion