Module 243
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.39 | 21 | 63 | In vitro growth | NA |
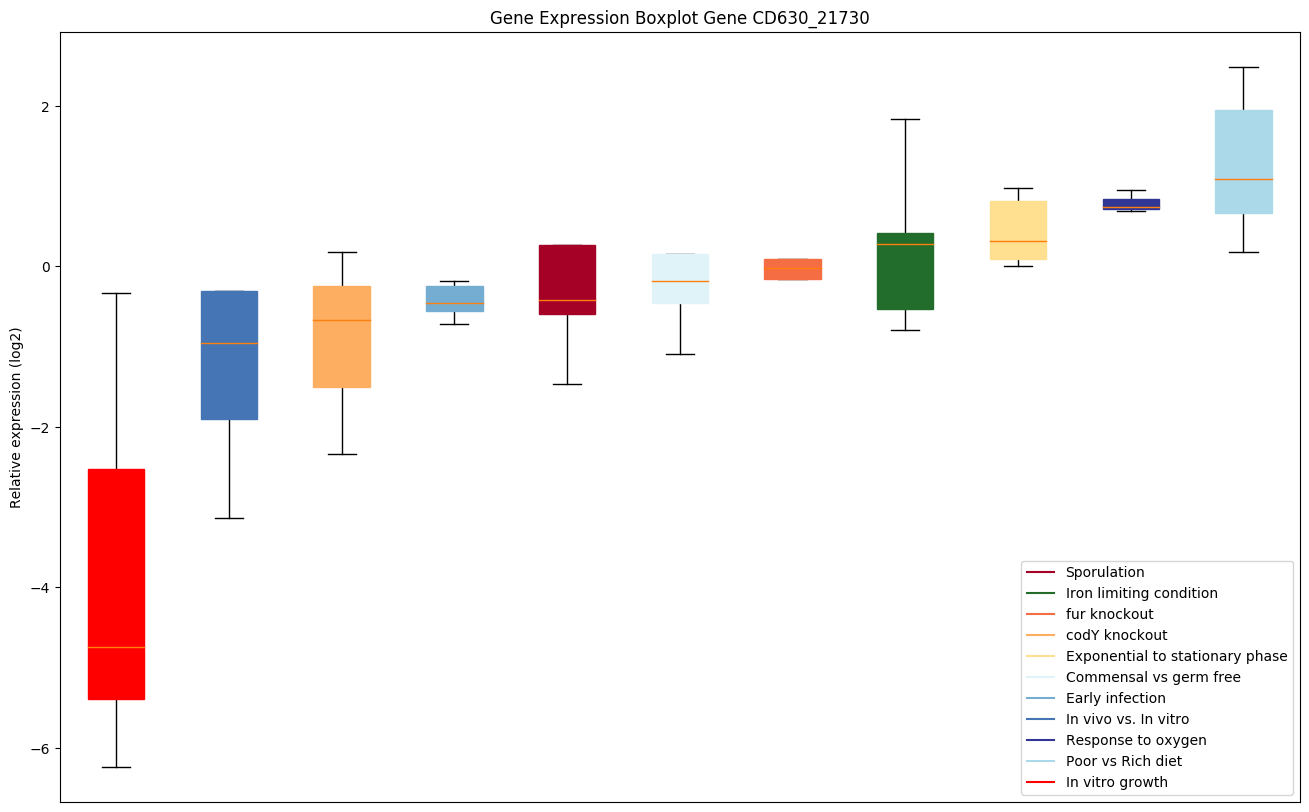
Bicluster expression profile
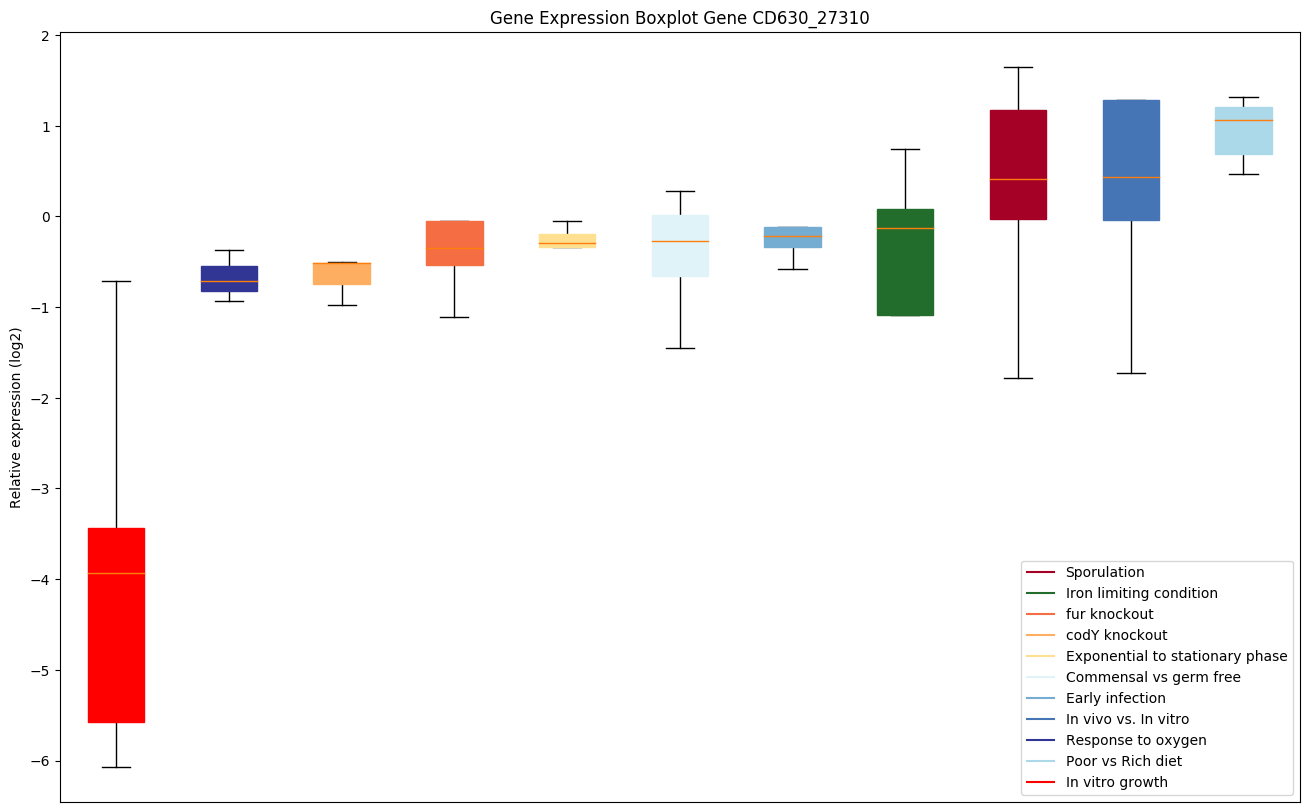
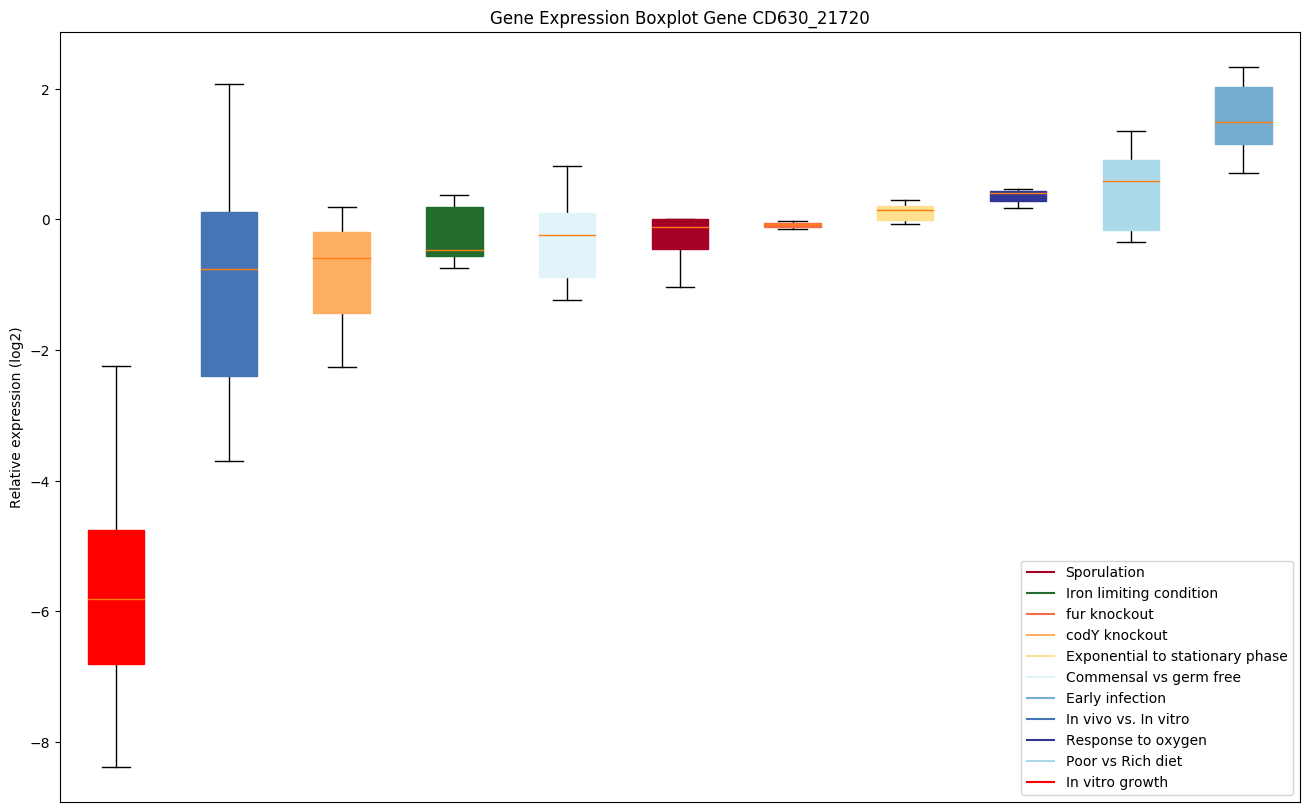
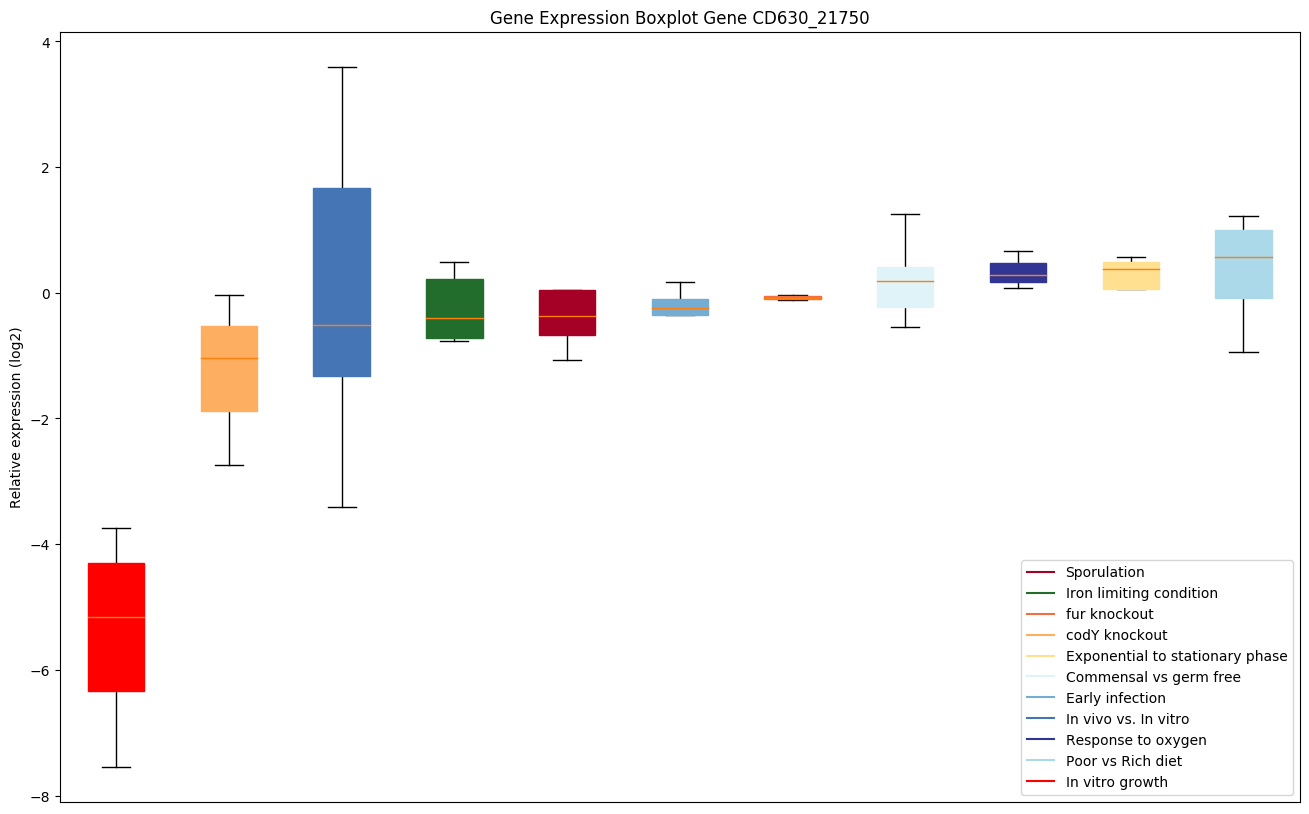
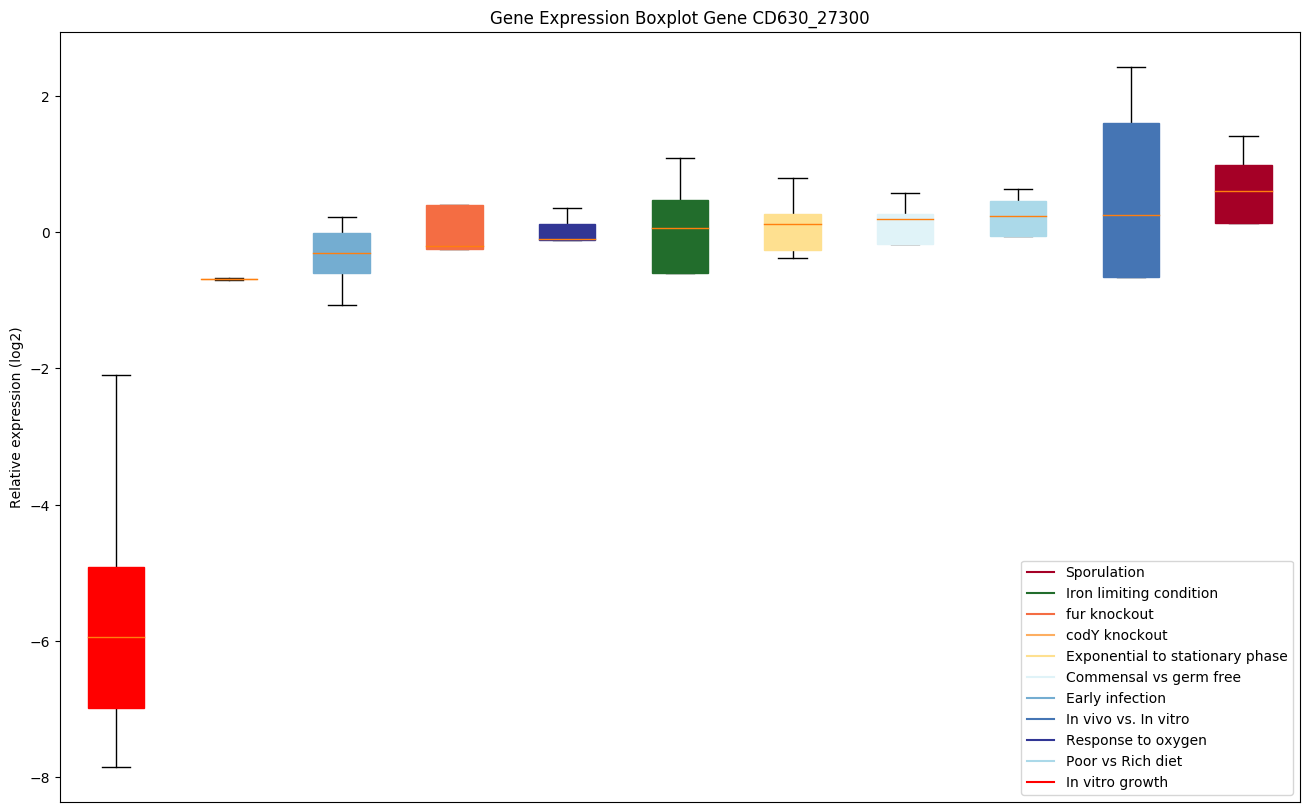
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
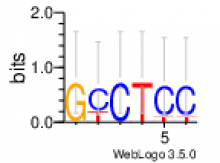
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
| TF | Module | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
|
prdR Transcriptional regulator, sigma-54-dependent |
243 | 6 | 0.00 | 0.01 |
The module is significantly enriched with genes associated to the indicated functional terms
The enrichment was determined with hypergeometric test using the genome functional annotation compiled by Girinathan et al (2020).
| Module | Pathway | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
| 243 | Transport Binding Proteins: Dipeptides and Oligopeptides | 6 | 0.000000 | 0.000000 |
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_24950 | selA | L-seryl-tRNA(Sec) selenium transferase(Selenocysteine synthase) (Sec synthase)(Selenocysteinyl-tRNA(Sec) synthase ) | Converts seryl-tRNA(Sec) to selenocysteinyl-tRNA(Sec) required for selenoprotein biosynthesis. |
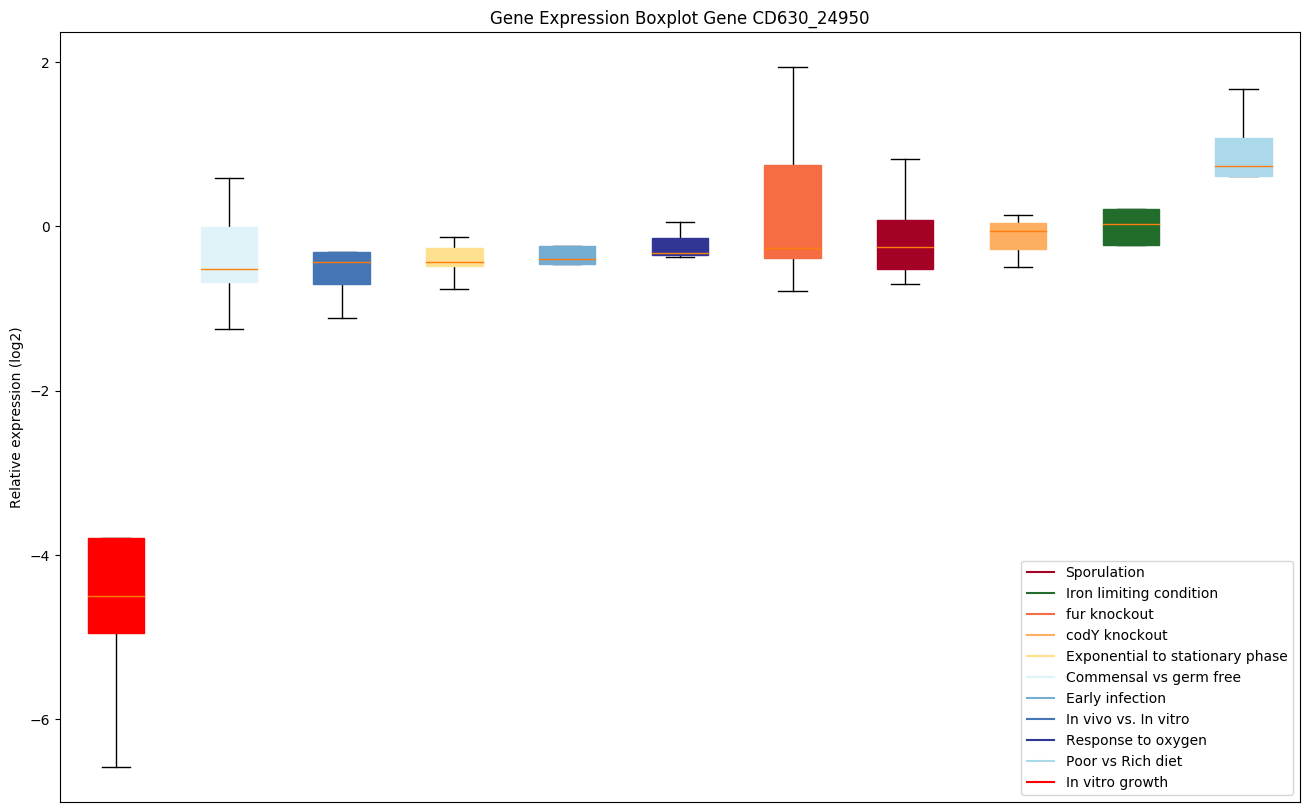 |
|||
| CD630_24980 | dacF | D-alanyl-D-alanine carboxypeptidase (putativesporulation-specific penicillin-binding protein) |
 |
||||
| CD630_24930 | selB | Selenocysteinyl-tRNA-specific translation factor |
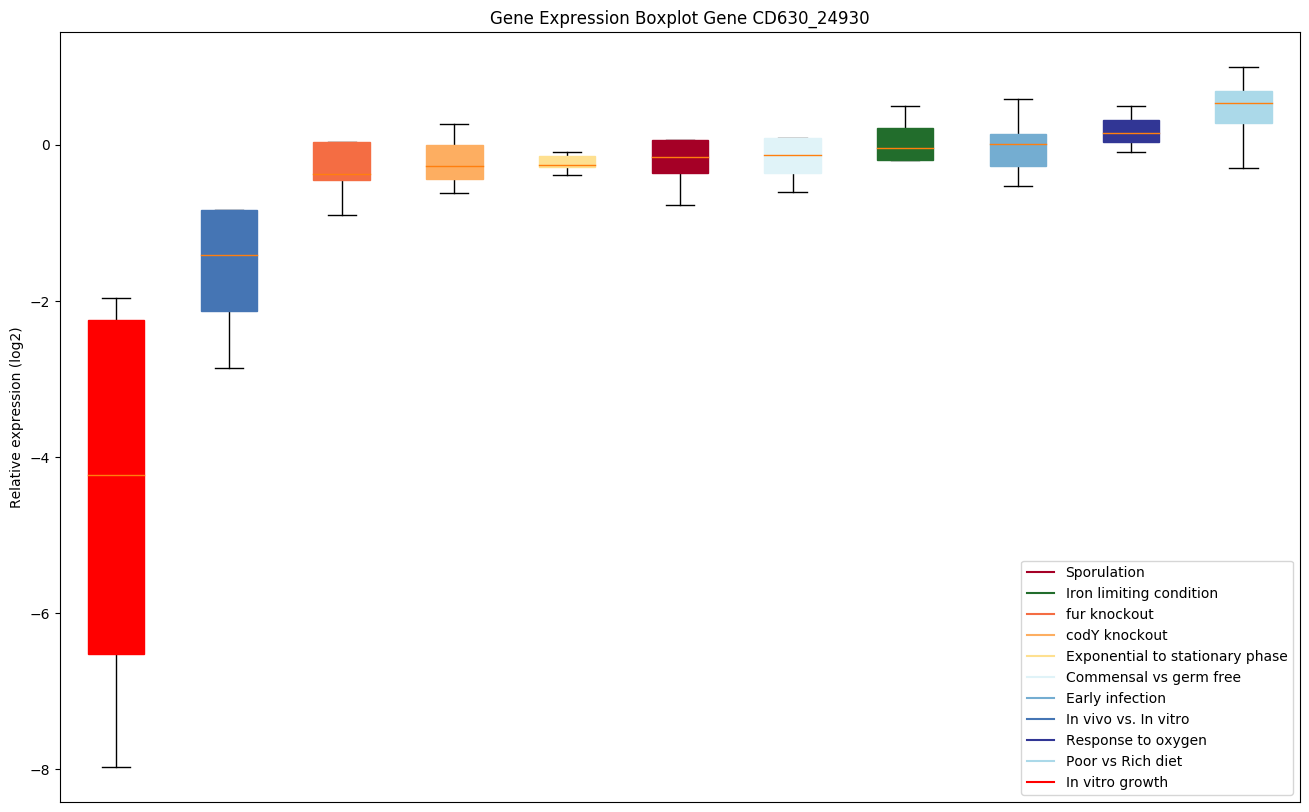 |
||||
| CD630_35140 | prs | Ribose-phosphate pyrophosphokinase | Involved in the biosynthesis of the central metabolite phospho-alpha-D-ribosyl-1-pyrophosphate (PRPP) via the transfer of pyrophosphoryl group from ATP to 1-hydroxyl of ribose-5-phosphate (Rib-5-P). | Yes | Yes | Yes |
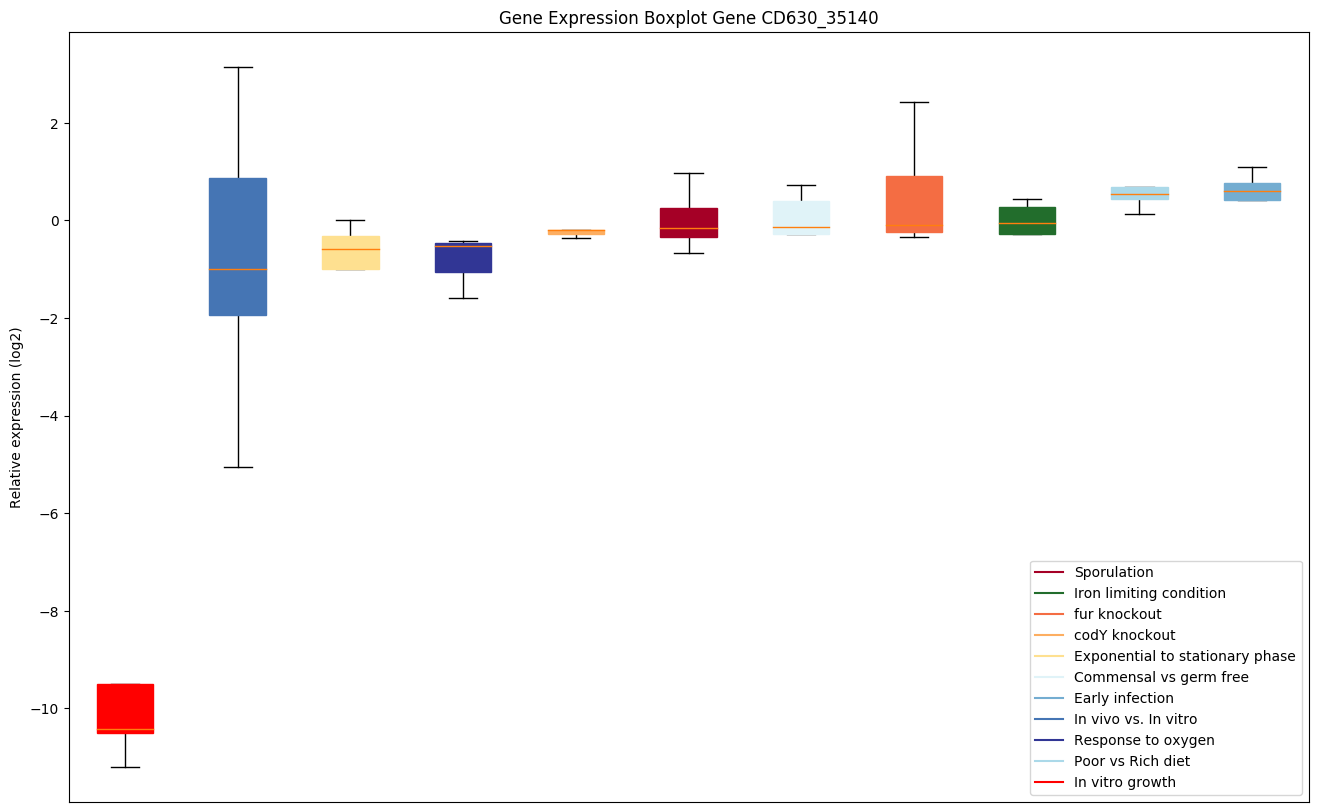 |
| CD630_01940 | groL | 60 kDa chaperonin (Protein Cpn60) (GroELprotein) | Prevents misfolding and promotes the refolding and proper assembly of unfolded polypeptides generated under stress conditions. | Yes |
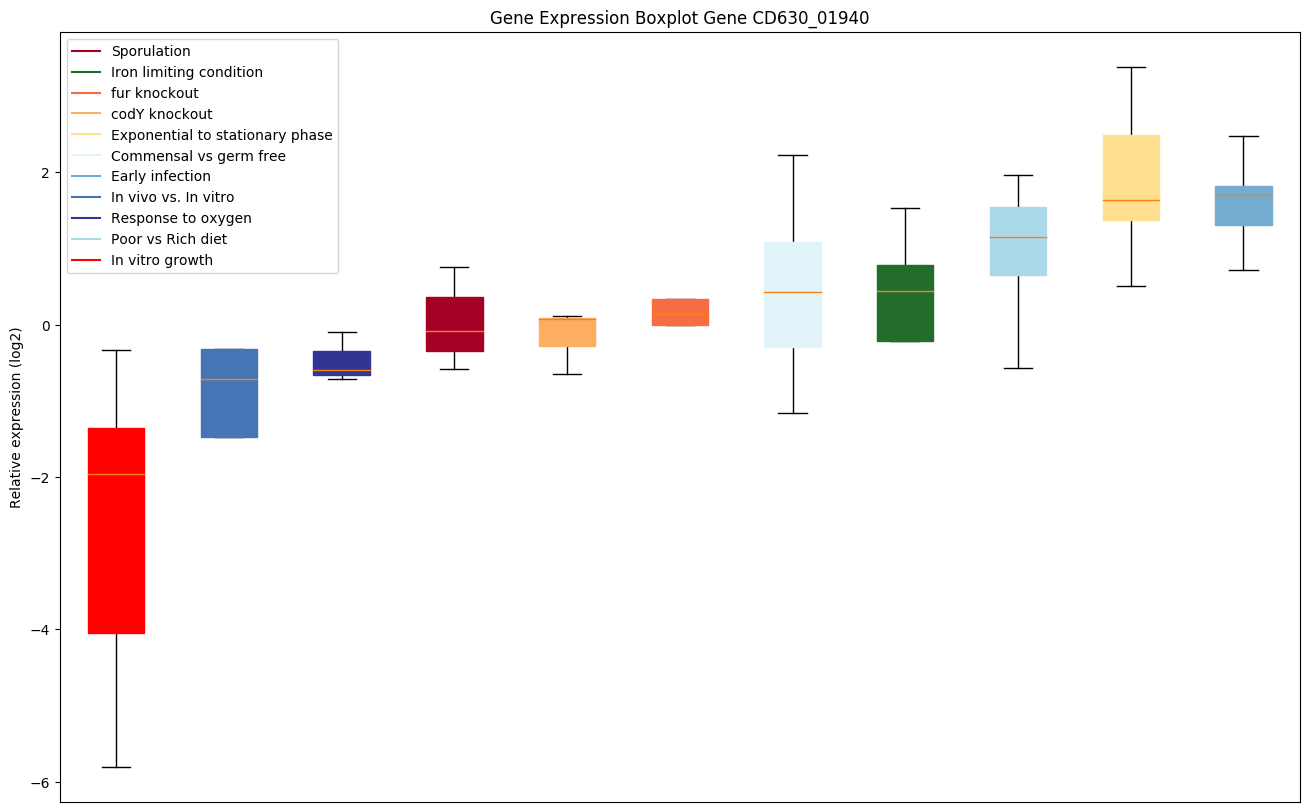 |
||
| CD630_25240 | nadD | Nicotinate-nucleotide adenylyltransferase | Catalyzes the reversible adenylation of nicotinate mononucleotide (NaMN) to nicotinic acid adenine dinucleotide (NaAD). | Yes |
 |
||
| CD630_24970 | comE | Competence protein ComEA |
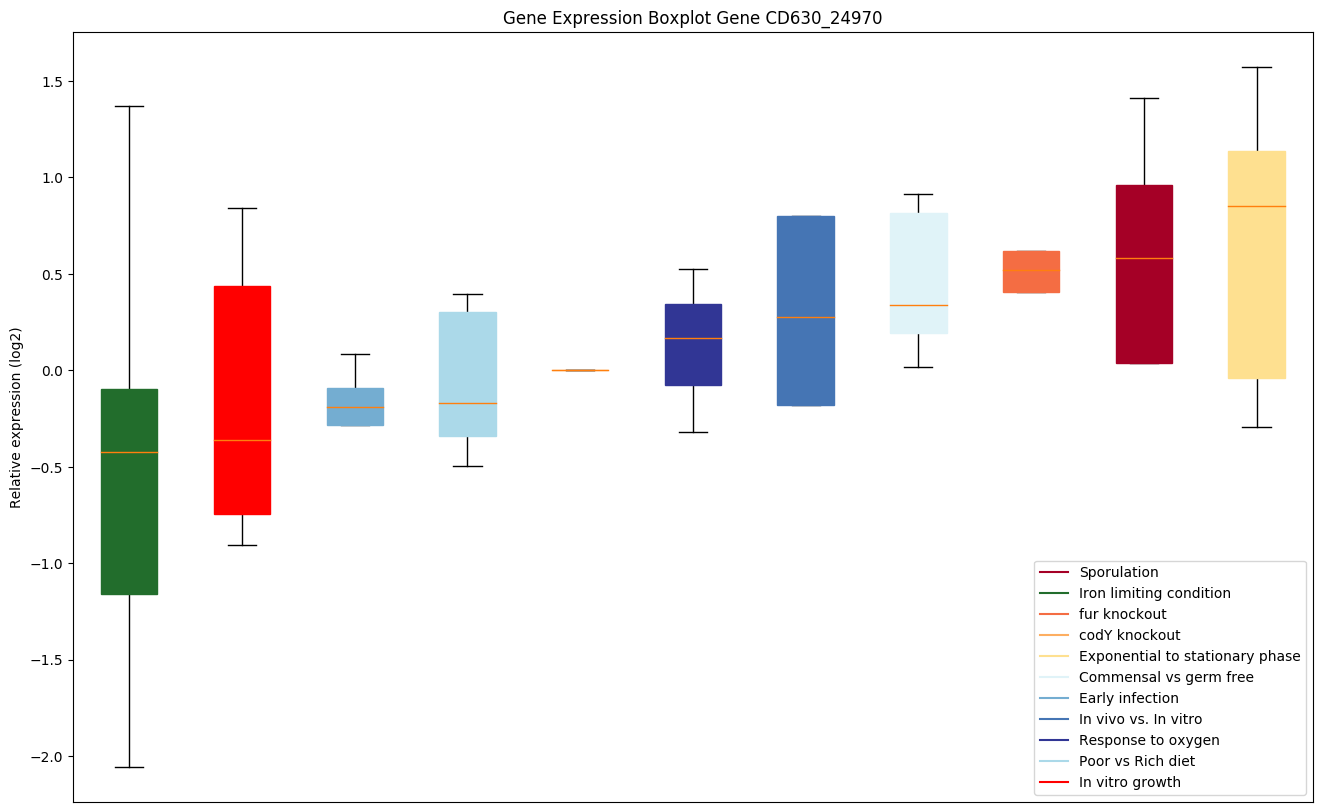 |
||||
| CD630_25230 | Putative hydrolase |
 |
|||||
| CD630_24960 | selD | Selenide, water dikinase (Selenophosphatesynthetase) (Selenium donor protein) | Synthesizes selenophosphate from selenide and ATP. |
 |
|||
| CD630_27290 | Putative regulator of the sigma(E) factor |
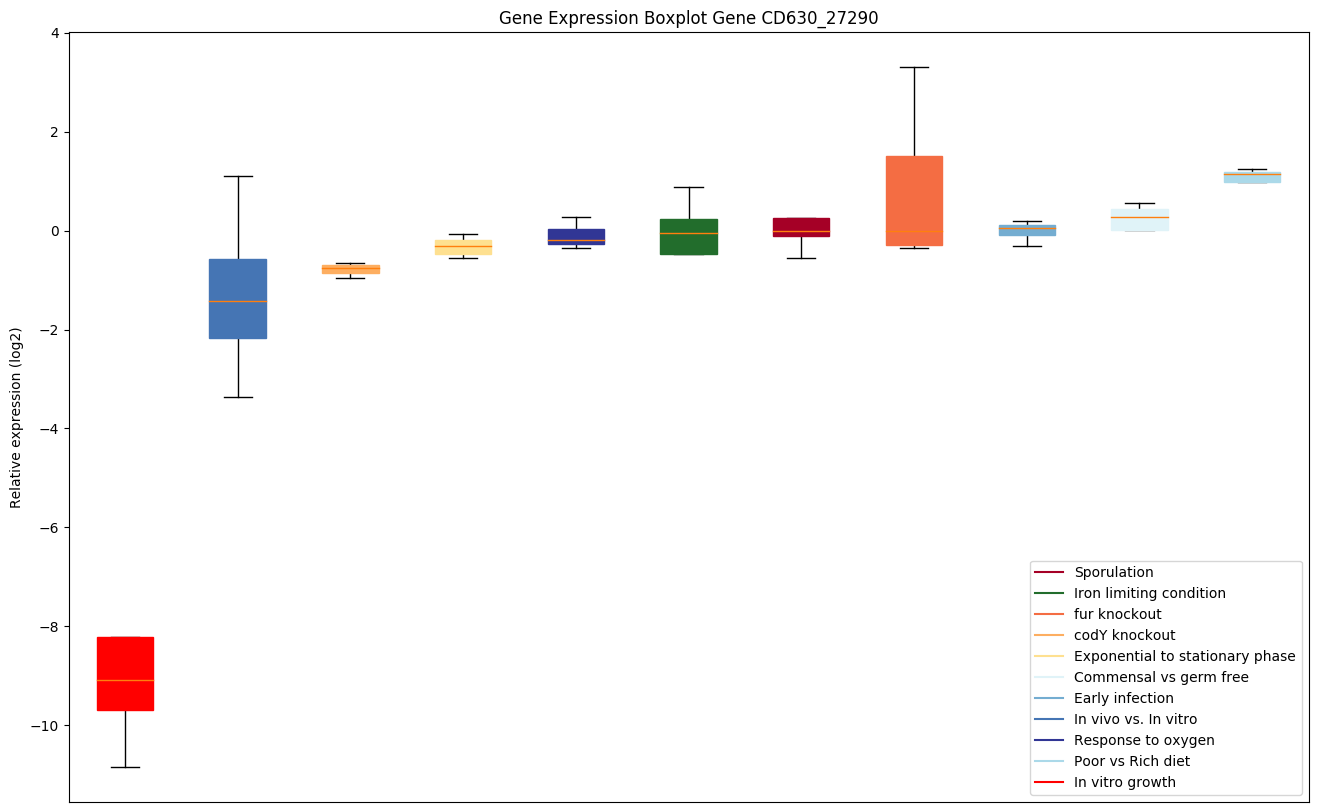 |
|||||
| CD630_01930 | groS | 10 kDa chaperonin (Protein Cpn10) (GroESprotein) | Binds to Cpn60 in the presence of Mg-ATP and suppresses the ATPase activity of the latter. | Yes |
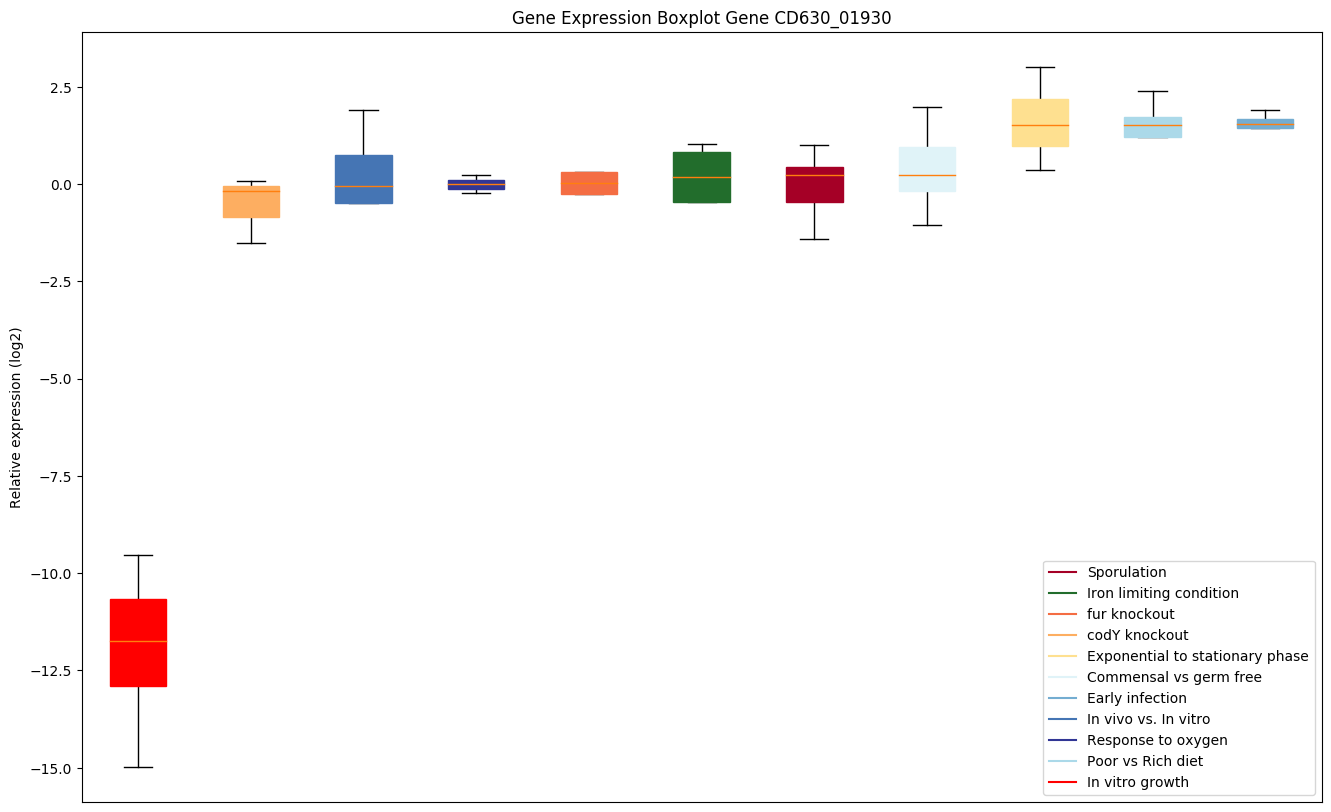 |
||
| CD630_21770 | ABC-type transport system,cystine/aminoacid-family extracellular solute-bindingprotein |
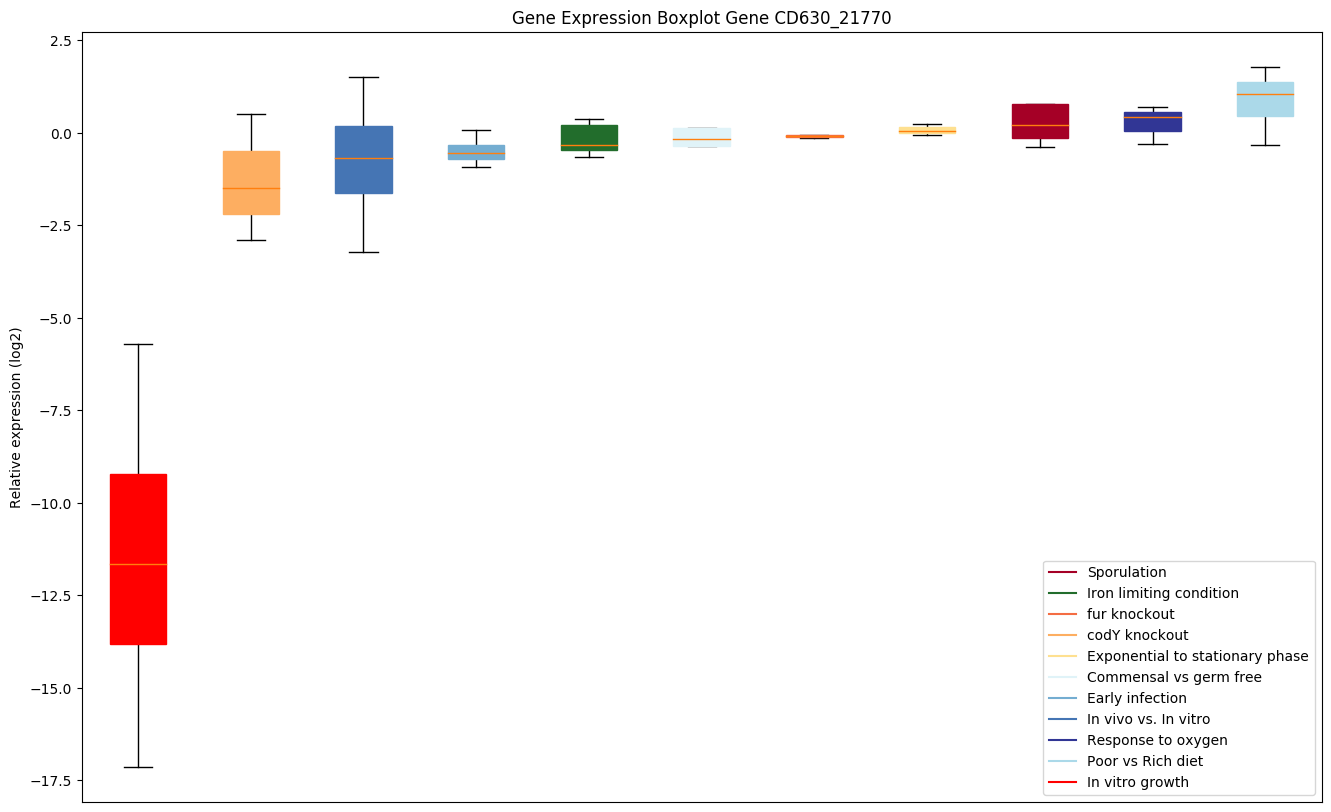 |
|||||
| CD630_25220 | Conserved hypothetical protein | Functions as a ribosomal silencing factor. Interacts with ribosomal protein L14 (rplN), blocking formation of intersubunit bridge B8. Prevents association of the 30S and 50S ribosomal subunits and the formation of functional ribosomes, thus repressing translation. |
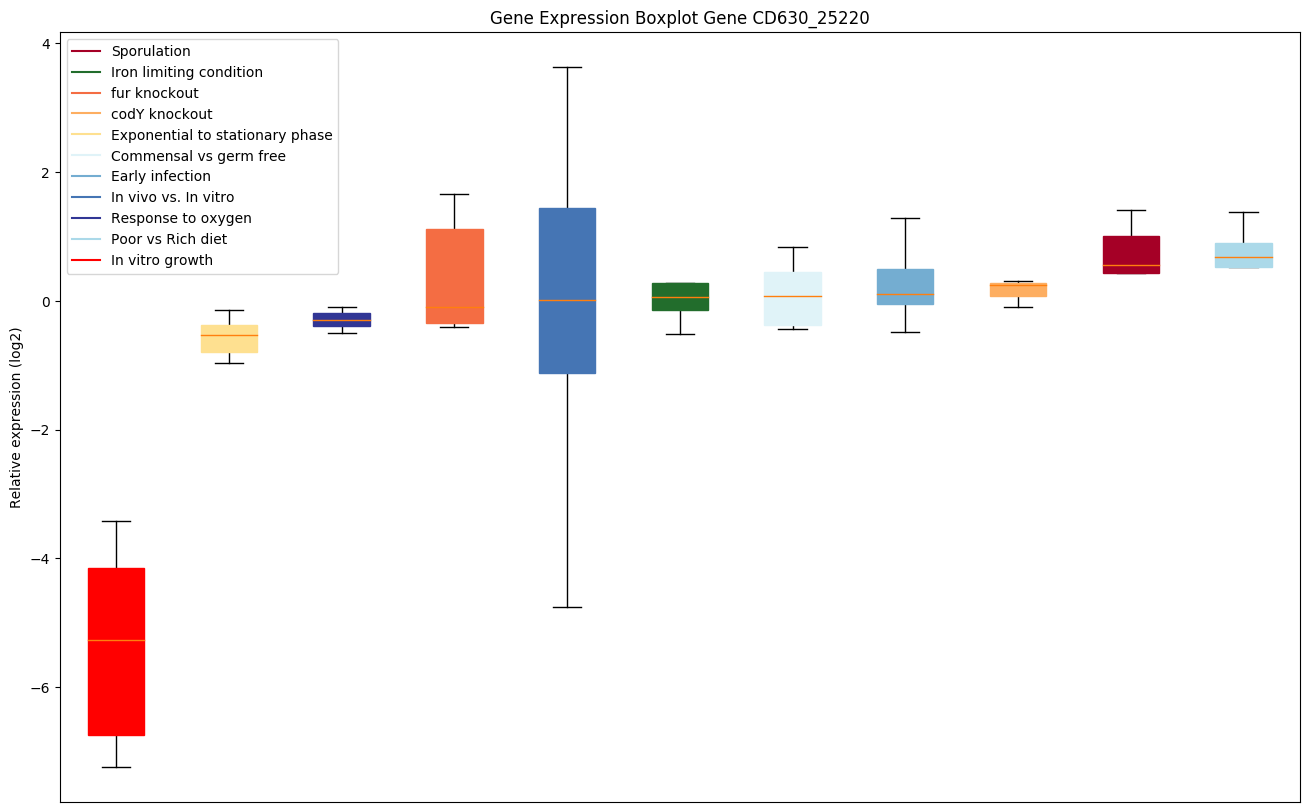 |
||||
| CD630_21760 | ABC-type transport system,cystine/aminoacid-family permease |
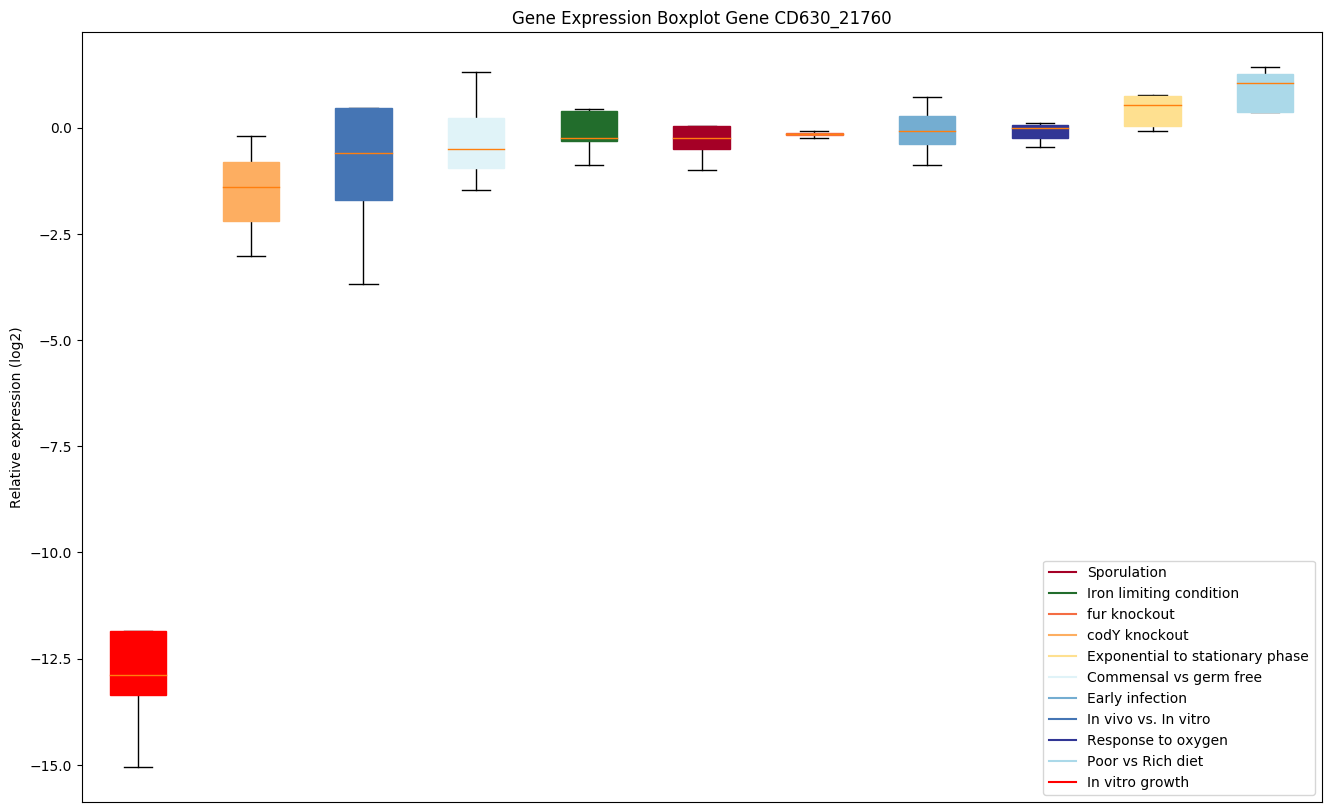 |
|||||
| CD630_35150 | glmU | Bifunctional protein GlmU [Includes:UDP-N-acetylglucosamine pyrophosphorylase ;Glucosamine-1-phosphate N-acetyltransferase] | Catalyzes the last two sequential reactions in the de novo biosynthetic pathway for UDP-N-acetylglucosamine (UDP-GlcNAc). The C-terminal domain catalyzes the transfer of acetyl group from acetyl coenzyme A to glucosamine-1-phosphate (GlcN-1-P) to produce N-acetylglucosamine-1-phosphate (GlcNAc-1-P), which is converted into UDP-GlcNAc by the transfer of uridine 5-monophosphate (from uridine 5-triphosphate), a reaction catalyzed by the N-terminal domain. | Yes | Yes | Yes |
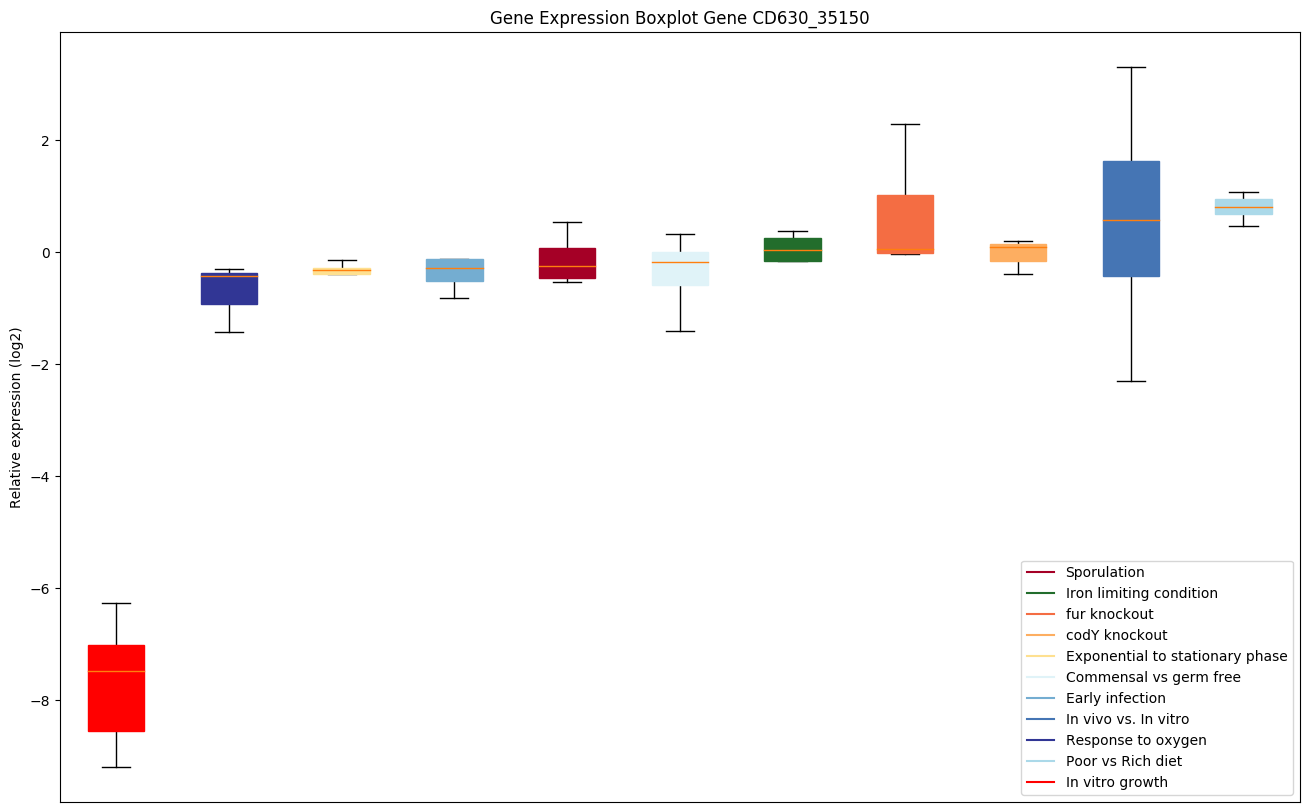 |
| CD630_27310 | Putative membrane protein |
 |
|||||
| CD630_21720 | ABC-type transport system,cystine/aminoacid-family ATP-binding protein |
 |
|||||
| CD630_21750 | ABC-type transport system,cystine/aminoacid-family permease |
 |
|||||
| CD630_21740 | ABC-type transport system,cystine/aminoacid-family extracellular solute-bindingprotein |
 |
|||||
| CD630_27300 | Conserved hypothetical protein |
 |
|||||
| CD630_21730 | Putative peptidase, M20D family |
 |