Module 333
Summary
| Residual | Gene Count | Condition Count | 1st Q Condition Block * | 5th Q Condition Block * |
|---|---|---|---|---|
| 0.54 | 18 | 63 | In vitro growth | NA |
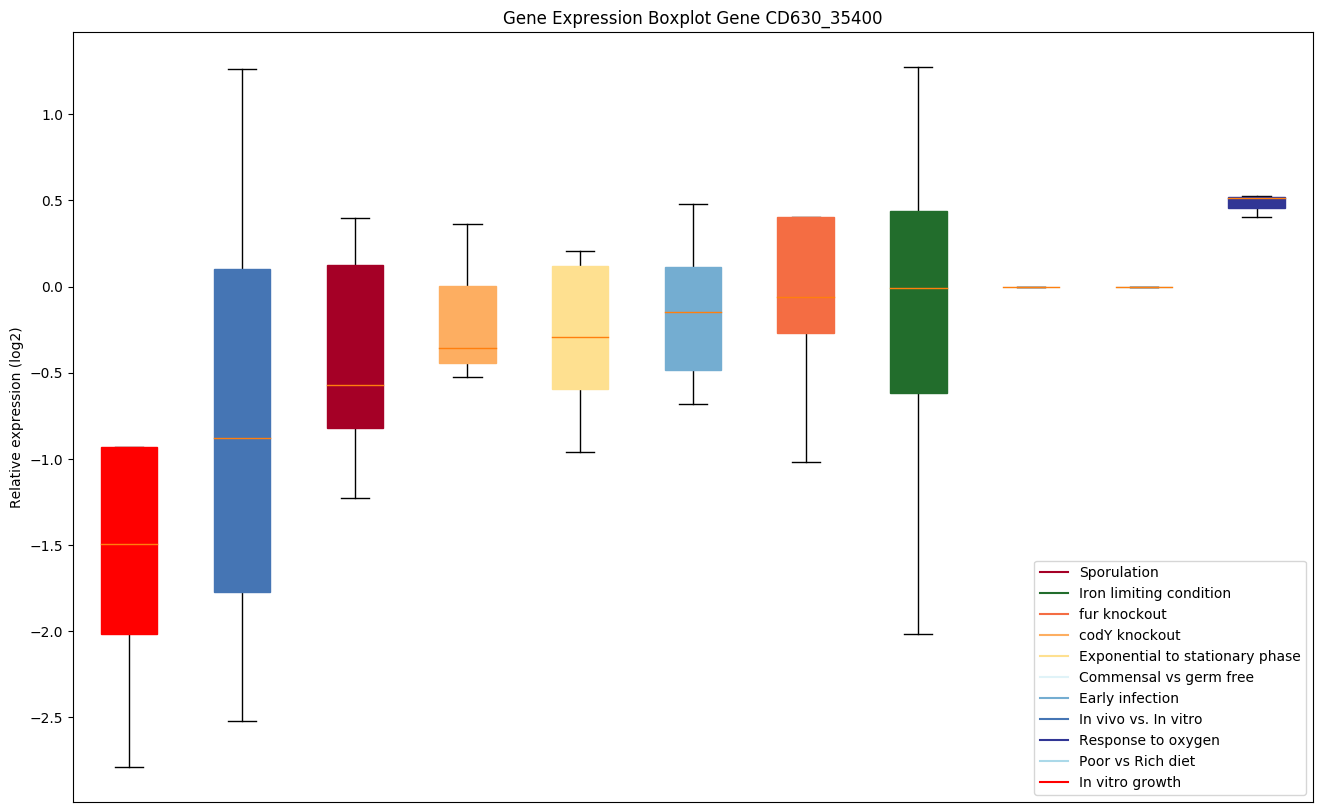
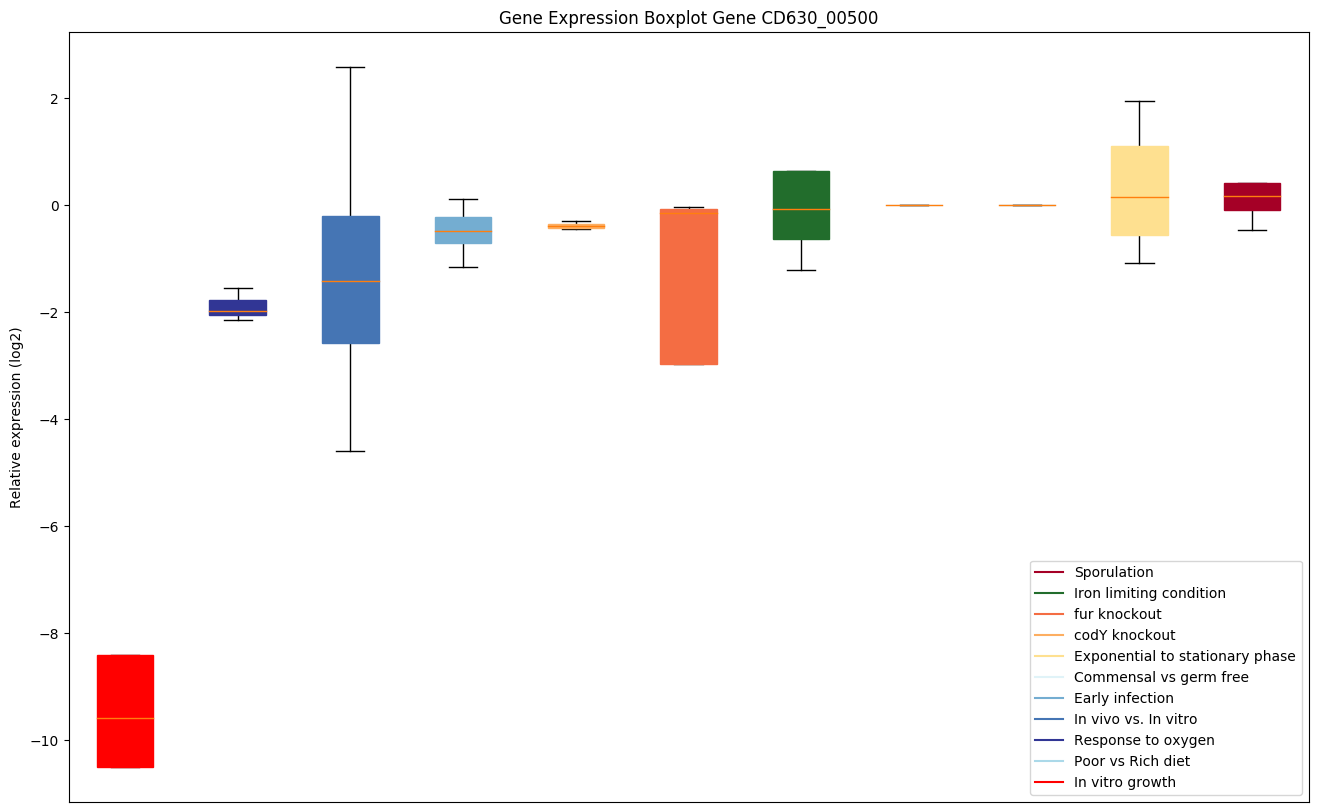
Bicluster expression profile
Expression of genes in subset of conditions included in bicluster on left side of red dashed line and out of bicluster on right of red dashed line. Each condition is represented as a boxplot, ordered by their median expression for the bicluster genes (smallest to largest) and colored according to condition blocks.
|
|
de novo identified motifs are listed below
Candidate transcriptional regulators for each module were determined with a linear regression based approach (Inferelator) and by evaluating the statistical significance of the overlap (Hypergeometric) between modules and putative TF and alternative sigma factor regulons (CcpA, CodY, Fur, PrdR, SigB, SigD, SigE, SigF, SigG, SigH, SigK, Spo0A) compiled from available literature.
For filtering high confidence influences, we used hypergeometric test adjusted p-value <=0.05 and minimum four genes in the overlap between the module and the TF regulon. The same thresholds were used for evaluating the functional enrichment.
1. Betas: correspond to the average coefficients of the Bayesian regressions between module expression and TF expression profiles. The values indicate the magnitude and direction (activation or repression for positive or negative values, respectively) of each TF-module interaction.
2. Confidence scores: indicate the likelihood of the TF-module interactions.
The module is significantly enriched with genes associated to the indicated functional terms
The enrichment was determined with hypergeometric test using the genome functional annotation compiled by Girinathan et al (2020).
| Module | Pathway | Overlap | pvalue | Adjusted pvalue |
|---|---|---|---|---|
| 333 | Amino-acyl tRNA Synthesis and Modification | 11 | 0.000000 | 0.000000 |
Genes that are included in this module
* "Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14".
| Title | Short Name | Product | Function | Essentiality * | in vivo Essentiality | Rich broth Essentiality | Expression |
|---|---|---|---|---|---|---|---|
| CD630_26180 | ileS | Isoleucyl-tRNA synthetase | Catalyzes the attachment of isoleucine to tRNA(Ile). As IleRS can inadvertently accommodate and process structurally similar amino acids such as valine, to avoid such errors it has two additional distinct tRNA(Ile)-dependent editing activities. One activity is designated as 'pretransfer' editing and involves the hydrolysis of activated Val-AMP. The other activity is designated 'posttransfer' editing and involves deacylation of mischarged Val-tRNA(Ile). | Yes |
 |
||
| CD630_12860 | Conserved hypothetical protein |
 |
|||||
| CD630_32560 | valS | valyl-tRNA synthetase | Catalyzes the attachment of valine to tRNA(Val). As ValRS can inadvertently accommodate and process structurally similar amino acids such as threonine, to avoid such errors, it has a 'posttransfer' editing activity that hydrolyzes mischarged Thr-tRNA(Val) in a tRNA-dependent manner. | Yes |
 |
||
| CD630_35400 | metG | Methionyl-tRNA synthetase (Methionine--tRNAligase) (MetRS) | Is required not only for elongation of protein synthesis but also for the initiation of all mRNA translation through initiator tRNA(fMet) aminoacylation. | Yes |
 |
||
| CD630_00500 | proS | Prolyl-tRNA synthetase | Catalyzes the attachment of proline to tRNA(Pro) in a two-step reaction: proline is first activated by ATP to form Pro-AMP and then transferred to the acceptor end of tRNA(Pro). | Yes |
 |
||
| CD630_26950 | asnA | Aspartate--ammonia ligase (Asparagine synthetaseA) |
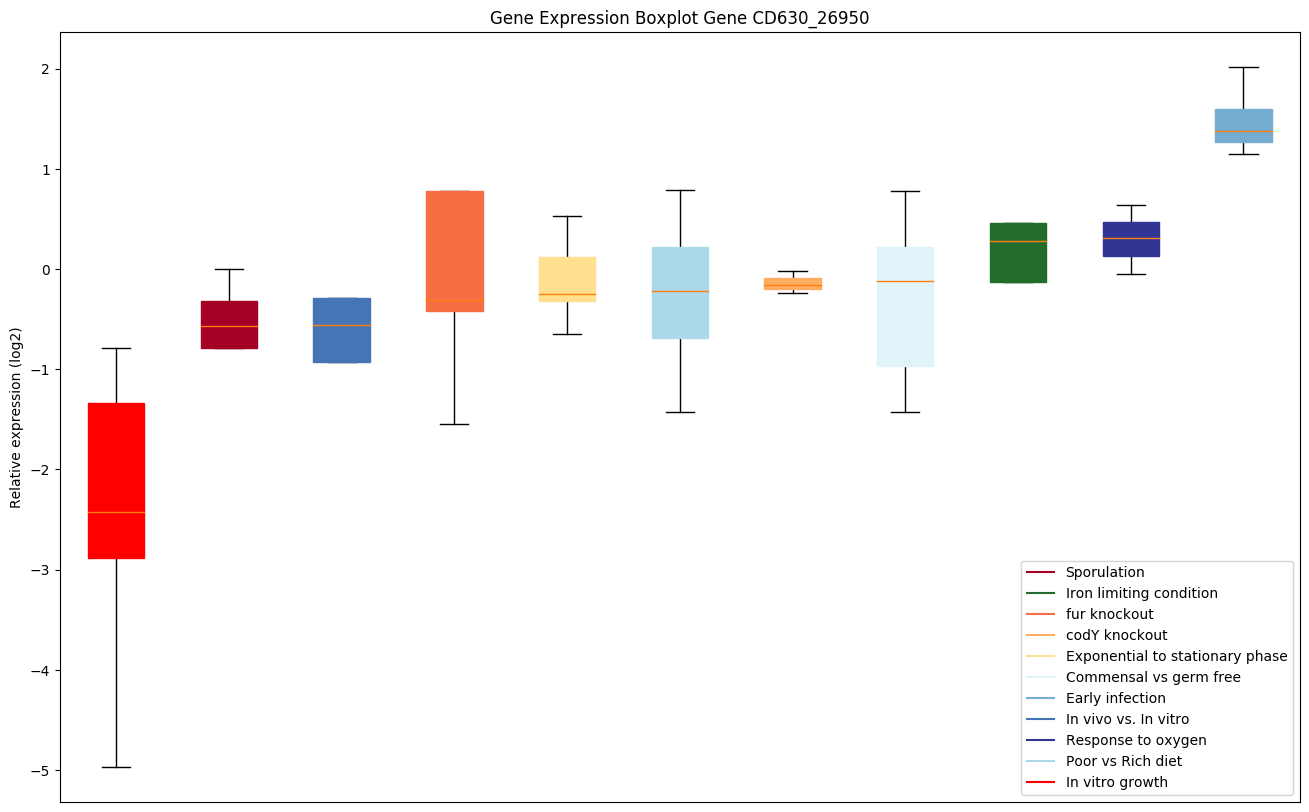 |
||||
| CD630_26940 | Conserved hypothetical protein |
 |
|||||
| CD630_26100 | trpS | Tryptophanyl-tRNA synthetase (Tryptophan--tRNAligase) (TrpRS) | Catalyzes the attachment of tryptophan to tRNA(Trp). | Yes |
 |
||
| CD630_26110 | mtnN | MTA/SAH nucleosidase (5'-methylthioadenosinenucleosidase) (S-adenosylhomocysteine nucleosidase) | Catalyzes the irreversible cleavage of the glycosidic bond in both 5'-methylthioadenosine (MTA) and S-adenosylhomocysteine (SAH/AdoHcy) to adenine and the corresponding thioribose, 5'-methylthioribose and S-ribosylhomocysteine, respectively. |
 |
|||
| CD630_35390 | Putative deoxyribonuclease |
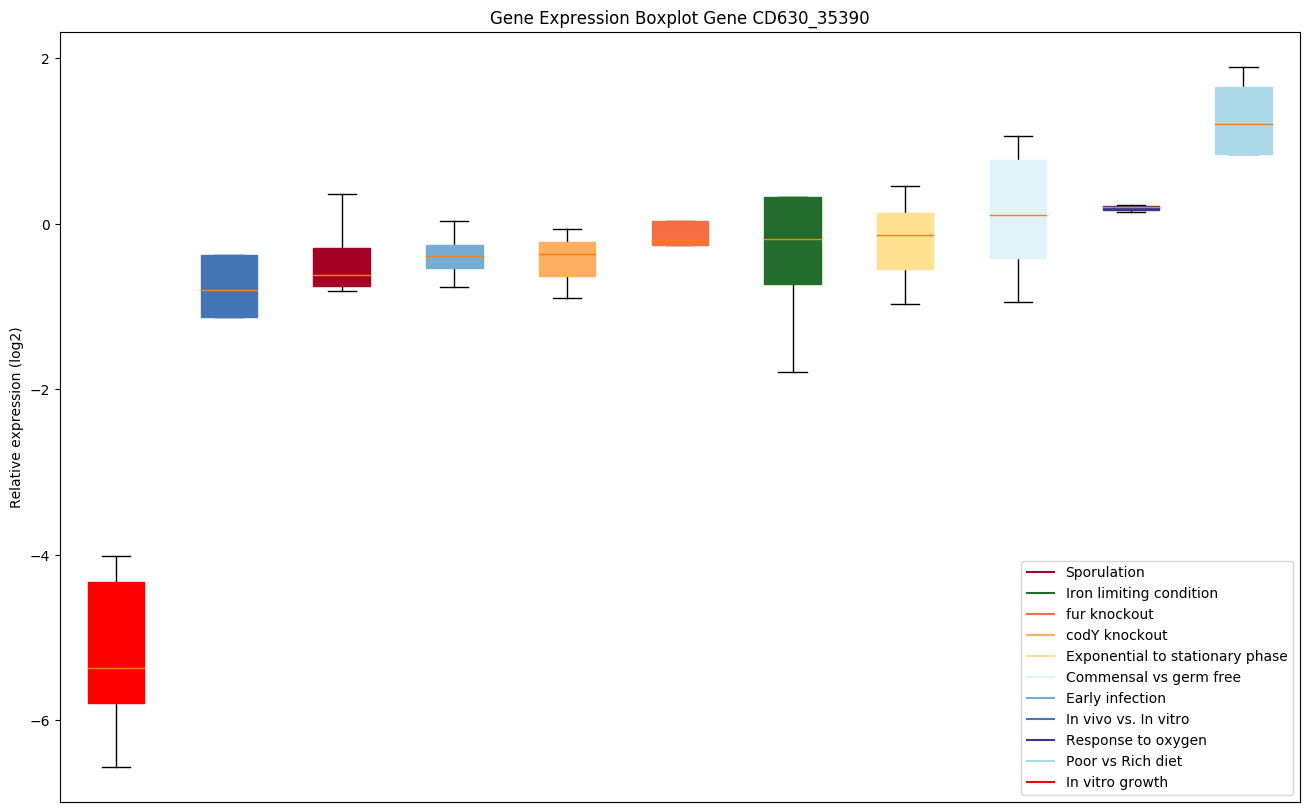 |
|||||
| CD630_00140 | serS1 | Seryl-tRNA synthetase 1 | Catalyzes the attachment of serine to tRNA(Ser). Is also able to aminoacylate tRNA(Sec) with serine, to form the misacylated tRNA L-seryl-tRNA(Sec), which will be further converted into selenocysteinyl-tRNA(Sec). | Yes |
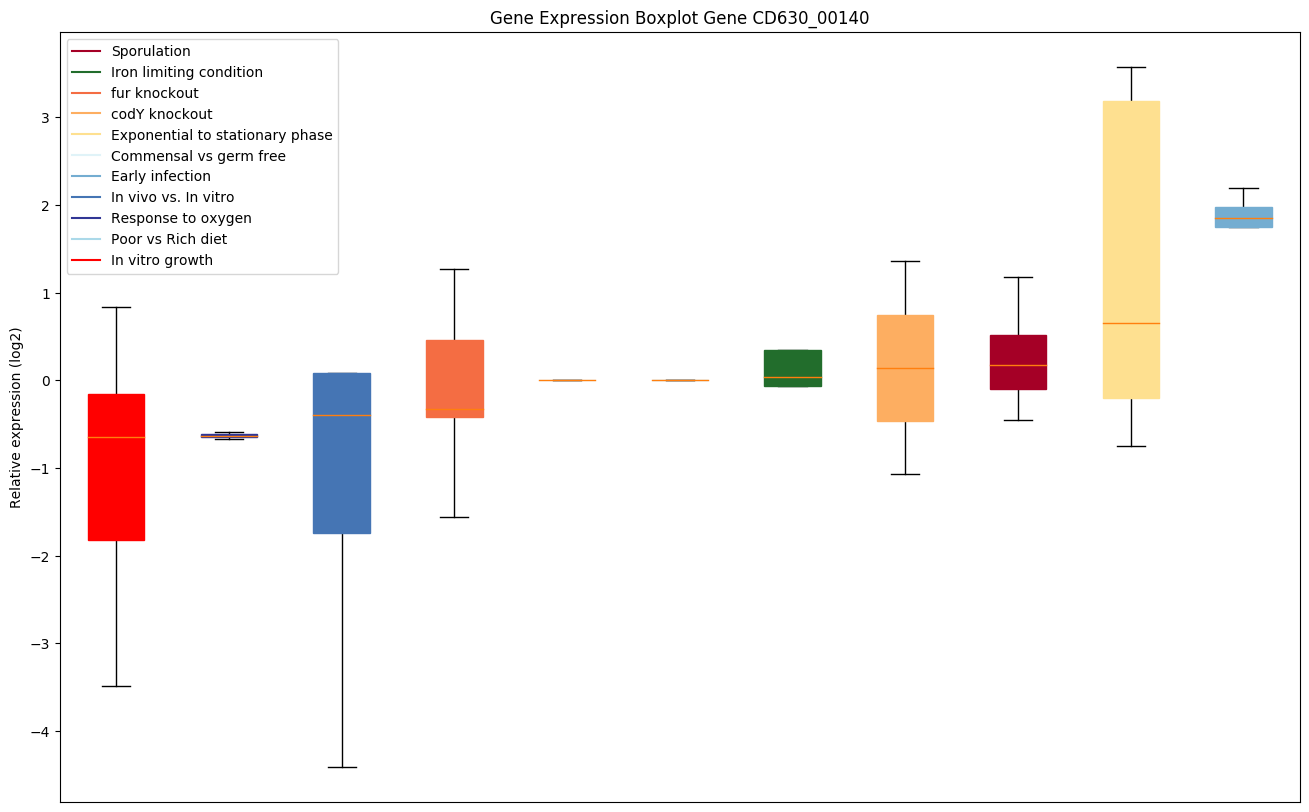 |
||
| CD630_00490 | proS | Prolyl-tRNA synthetase | Catalyzes the attachment of proline to tRNA(Pro) in a two-step reaction: proline is first activated by ATP to form Pro-AMP and then transferred to the acceptor end of tRNA(Pro). As ProRS can inadvertently accommodate and process non-cognate amino acids such as alanine and cysteine, to avoid such errors it has two additional distinct editing activities against alanine. One activity is designated as 'pretransfer' editing and involves the tRNA(Pro)-independent hydrolysis of activated Ala-AMP. The other activity is designated 'posttransfer' editing and involves deacylation of mischarged Ala-tRNA(Pro). The misacylated Cys-tRNA(Pro) is not edited by ProRS. | Yes |
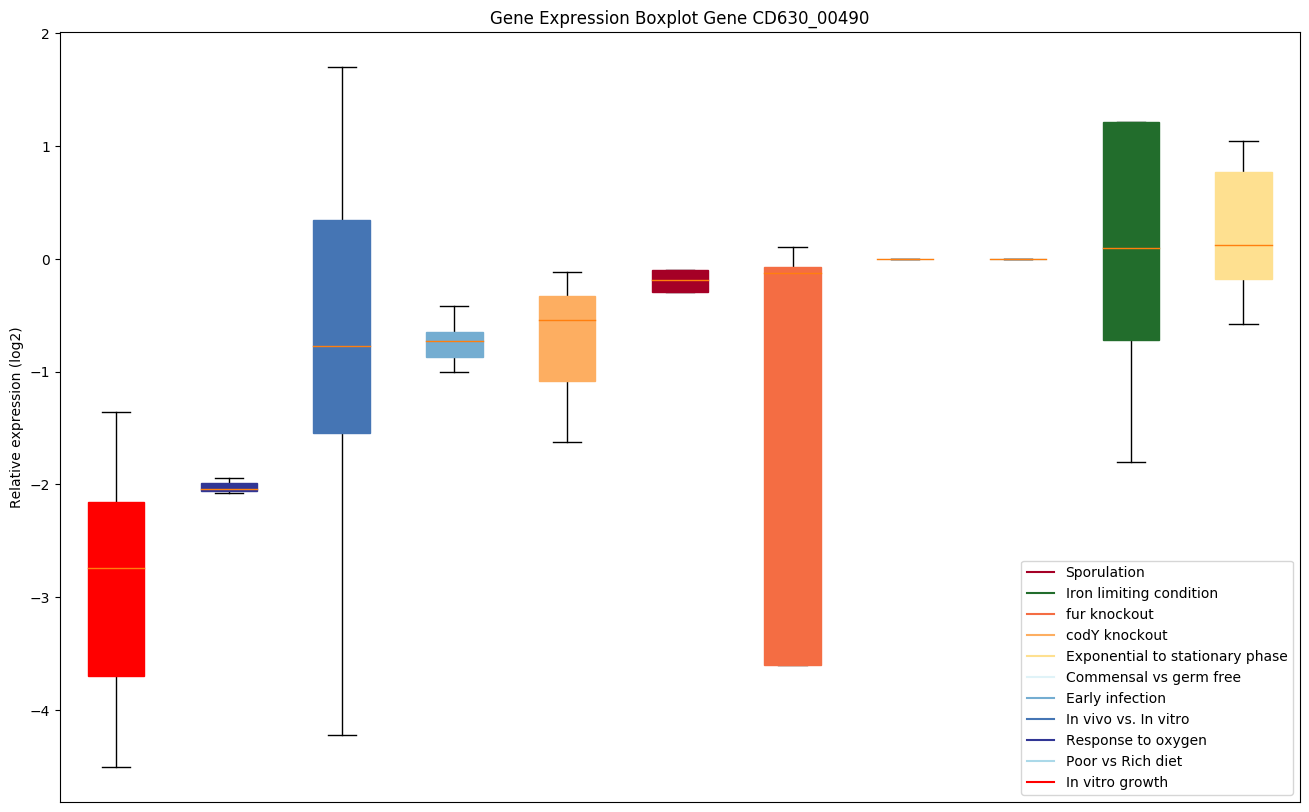 |
||
| CD630_25210 | leuS | Leucyl-tRNA synthetase | Yes |
 |
|||
| CD630_07010 | Putative cell-division protein | Activator of cell division through the inhibition of FtsZ GTPase activity, therefore promoting FtsZ assembly into bundles of protofilaments necessary for the formation of the division Z ring. It is recruited early at mid-cell but it is not essential for cell division. | Yes |
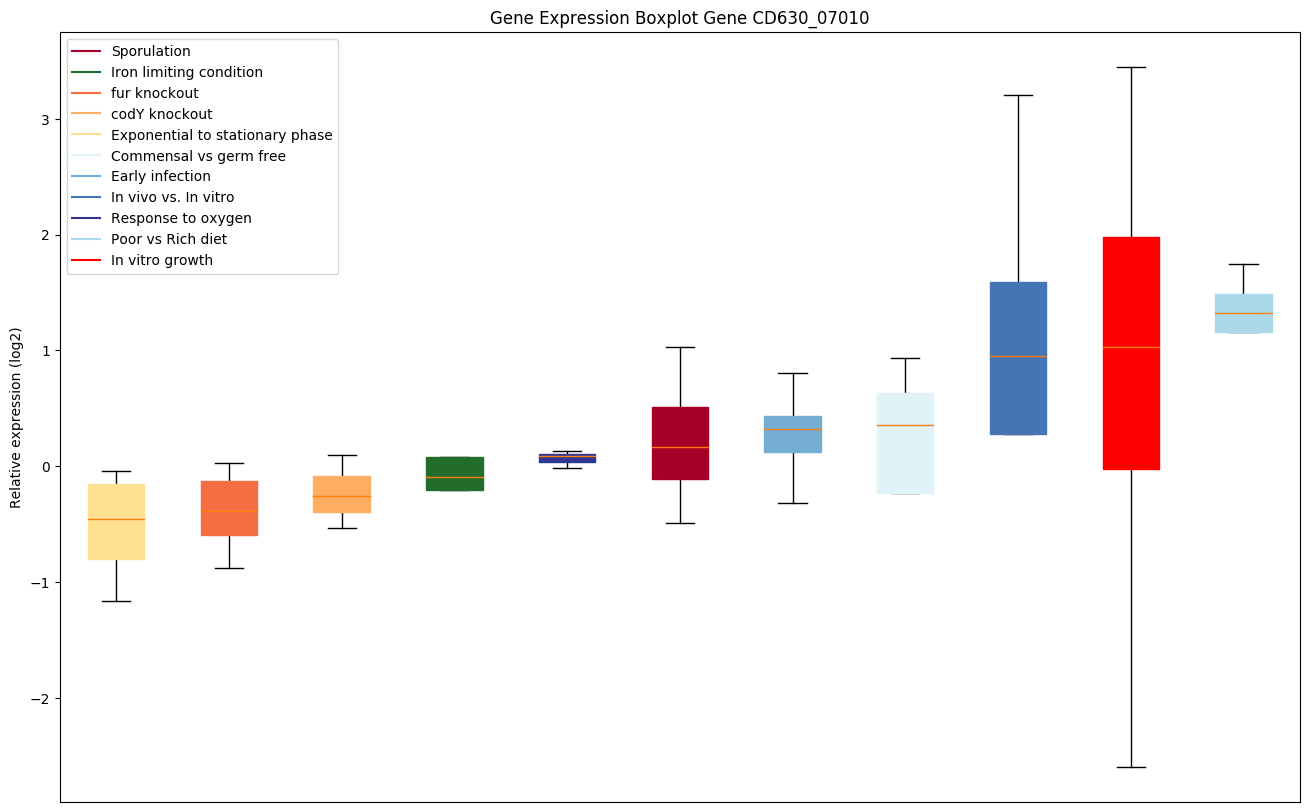 |
|||
| CD630_12850 | Putative Holliday junction resolvase | Could be a nuclease involved in processing of the 5'-end of pre-16S rRNA. |
 |
||||
| CD630_07000 | pheT | Phenylalanyl-tRNA synthetase subunit beta(Phenylalanine--tRNA ligase beta chain) (PheRS) | Yes |
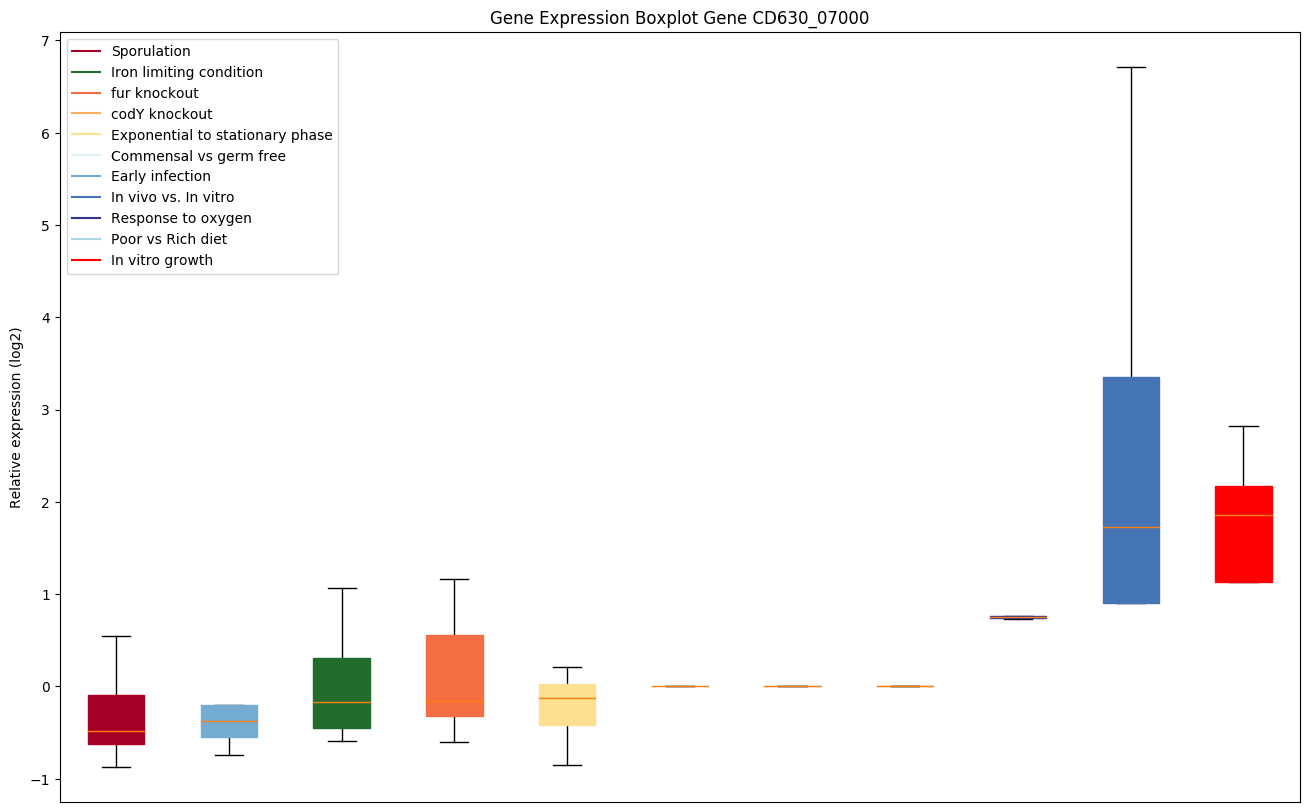 |
|||
| CD630_15210 | tyrS | Tyrosyl-tRNA synthetase (Tyrosine--tRNA ligase)(TyrRS) | Catalyzes the attachment of tyrosine to tRNA(Tyr) in a two-step reaction: tyrosine is first activated by ATP to form Tyr-AMP and then transferred to the acceptor end of tRNA(Tyr). | Yes |
 |
||
| CD630_12820 | alaS | Alanyl-tRNA synthetase | Catalyzes the attachment of alanine to tRNA(Ala) in a two-step reaction: alanine is first activated by ATP to form Ala-AMP and then transferred to the acceptor end of tRNA(Ala). Also edits incorrectly charged Ser-tRNA(Ala) and Gly-tRNA(Ala) via its editing domain. | Yes |
 |