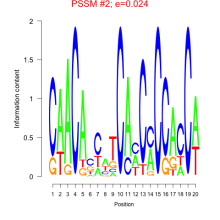
Thaps_bicluster_0152 Residual: 0.38
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0152 | 0.38 | Thalassiosira pseudonana |
Displaying 1 - 32 of 32
" class="views-fluidgrid-wrapper clear-block">
10379 hypothetical protein
10475 hypothetical protein
10564 DadA
11341 AAT_like
1382 GAT_1 superfamily
15819 Arginase_HDAC superfamily
16039 MutS
20619 hypothetical protein
21308 RING
21827 hypothetical protein
2310 hypothetical protein
2451 PH-like superfamily
25325 hypothetical protein
25565 hypothetical protein
263404 Peptidase_C1 superfamily
264145 HA
268652 ATG11 superfamily
268823 Aa_trans superfamily
2773 hypothetical protein
28413 BglB superfamily
31052 S_TKc
32899 HlyIII superfamily
33717 PLN02856
4482 hypothetical protein
5051 hypothetical protein
5820 hypothetical protein
6818 AAT_I superfamily
6826 PH-like superfamily
7980 Sulfotransfer_1
8445 hypothetical protein
9018 RdRP
9410 Transglut_core
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009072 | aromatic amino acid family metabolism | 0.00619065 | 1 | Thaps_bicluster_0152 |
| GO:0006298 | mismatch repair | 0.0326202 | 1 | Thaps_bicluster_0152 |
| GO:0006865 | amino acid transport | 0.0584155 | 1 | Thaps_bicluster_0152 |
| GO:0009058 | biosynthesis | 0.108157 | 1 | Thaps_bicluster_0152 |
| GO:0006508 | proteolysis and peptidolysis | 0.115922 | 1 | Thaps_bicluster_0152 |
| GO:0005975 | carbohydrate metabolism | 0.144797 | 1 | Thaps_bicluster_0152 |
| GO:0016567 | protein ubiquitination | 0.15196 | 1 | Thaps_bicluster_0152 |
| GO:0006468 | protein amino acid phosphorylation | 0.362866 | 1 | Thaps_bicluster_0152 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.481263 | 1 | Thaps_bicluster_0152 |

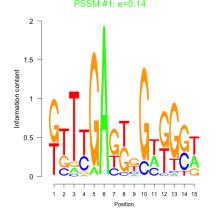
Comments