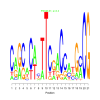
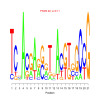
Module 12 Residual: 0.58
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0012 | v02 | 0.58 | -19.63 |
| Rv0088 | Rv0125 Serine protease PepA |
| Rv0131c PROBABLE ACYL-CoA DEHYDROGENASE FADE1 (EC 1.3.99.-) | Rv0308 Possible membrane protein |
| Rv1142c Enoyl-CoA hydratase (EC 4.2.1.17) | Rv1482c |
| Rv1483 3-oxoacyl-[acyl-carrier protein] reductase (EC 1.1.1.100) | Rv1484 Enoyl-[acyl-carrier-protein] reductase [NADH] (EC 1.3.1.9) |
| Rv1485 Ferrochelatase, protoheme ferro-lyase (EC 4.99.1.1) | Rv2280 Probable dehydrogenase |
| Rv2998 | Rv3000 YbbM seven transmembrane helix protein |
| Rv3220c Signal transduction histidine kinase, subgroup 2 |
|
GO:0005618 | cell wall |
|
GO:0005886 | plasma membrane |
|
GO:0005576 | extracellular region |
|
GO:0005829 | cytosol |
|
GO:0004300 | enoyl-CoA hydratase activity |
|
GO:0004316 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) activity |
|
GO:0018454 | acetoacetyl-CoA reductase activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0051289 | protein homotetramerization |
|
GO:0071768 | mycolic acid biosynthetic process |
|
GO:0004318 | enoyl-[acyl-carrier-protein] reductase (NADH) activity |
|
GO:0005504 | fatty acid binding |
|
GO:0030497 | fatty acid elongation |
|
GO:0070403 | NAD+ binding |
|
GO:0004325 | ferrochelatase activity |
|
GO:0006783 | heme biosynthetic process |
|
GO:0040007 | growth |
|
GO:0051537 | 2 iron, 2 sulfur cluster binding |
|
GO:0000155 | two-component sensor activity |
|
GO:0000160 | two-component signal transduction system (phosphorelay) |
|
GO:0004672 | protein kinase activity |
|
GO:0005524 | ATP binding |
|
GO:0046777 | protein autophosphorylation |















Discussion