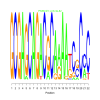
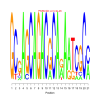
Module 98 Residual: 0.64
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0098 | v02 | 0.64 | -19.60 |
| Rv0153c Protein tyrosine phosphatase (EC 3.1.3.48) | Rv0154c Butyryl-CoA dehydrogenase (EC 1.3.99.2) |
| Rv0686 FIG025093: Probable membrane protein | Rv0687 3-oxoacyl-[acyl-carrier protein] reductase (EC 1.1.1.100) |
| Rv0688 Ferredoxin reductase | Rv0712 Sulfatase modifying factor 1 precursor (C-alpha-formyglycine- generating enzyme 1) |
| Rv0724 Possible protease IV SppA (endopeptidase IV) (signal peptide peptidase) | Rv0902c Sensor-type histidine kinase prrB (EC 2.7.13.3) |
| Rv0903c DNA-binding response regulator | Rv1059 |
| Rv1060 | Rv1096 Possible glycosyl hydrolase (EC 3.-.-.-) |
| Rv1179c DNA or RNA helicases of superfamily II | Rv1313c Mobile element protein |
| Rv1831 | Rv2061c |
| Rv2188c Poly(glycerol-phosphate) alpha-glucosyltransferase (EC 2.4.1.52) | Rv2248 |
| Rv3247c Thymidylate kinase (EC 2.7.4.9) | Rv3491 |
|
GO:0004437 | inositol or phosphatidylinositol phosphatase activity |
|
GO:0004722 | protein serine/threonine phosphatase activity |
|
GO:0004725 | protein tyrosine phosphatase activity |
|
GO:0005576 | extracellular region |
|
GO:0006470 | protein dephosphorylation |
|
GO:0009405 | pathogenesis |
|
GO:0004438 | phosphatidylinositol-3-phosphatase activity |
|
GO:0005886 | plasma membrane |
|
GO:0005515 | protein binding |
|
GO:0005887 | integral to plasma membrane |
|
GO:0008860 | ferredoxin-NAD+ reductase activity |
|
GO:0009055 | electron carrier activity |
|
GO:0050660 | flavin adenine dinucleotide binding |
|
GO:0051287 | NAD binding |
|
GO:0018083 | peptidyl-L-3-oxoalanine biosynthetic process from peptidyl-cysteine or peptidyl-serine |
|
GO:0004672 | protein kinase activity |
|
GO:0005524 | ATP binding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0044110 | growth involved in symbiotic interaction |
|
GO:0046777 | protein autophosphorylation |
|
GO:0008150 | biological_process |
|
GO:0000287 | magnesium ion binding |
|
GO:0005509 | calcium ion binding |
|
GO:0005829 | cytosol |
|
GO:0052572 | response to host immune response |
|
GO:0005618 | cell wall |
|
GO:0000030 | mannosyltransferase activity |
|
GO:0009247 | glycolipid biosynthetic process |
|
GO:0040007 | growth |
|
GO:0004798 | thymidylate kinase activity |
|
GO:0005525 | GTP binding |
|
GO:0046044 | TMP metabolic process |






















Discussion