Module 164 Residual: 0.60
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0164 | v02 | 0.60 | -34.88 |
| Rv0427c Exodeoxyribonuclease III (EC 3.1.11.2) | Rv0429c Peptide deformylase (EC 3.5.1.88) |
| Rv0503c Cyclopropane-fatty-acyl-phospholipid synthase 2, CmaA2 (EC 2.1.1.79) | Rv0504c (3R)-hydroxyacyl-ACP dehydratase subunit HadA |
| Rv0994 Molybdopterin biosynthesis protein MoeA | Rv1668c |
| Rv1822 CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase (EC 2.7.8.5) | Rv1826 Glycine cleavage system H protein |
| Rv1867 Acetyl-CoA acetyltransferase | Rv2113 Possible membrane protein |
| Rv2141c FIG016551: Putative peptidase | Rv3302c Glycerol-3-phosphate dehydrogenase (EC 1.1.5.3) |
| Rv3303c Dihydrolipoamide dehydrogenase (EC 1.8.1.4) | Rv3668c Protease |
| Rv3669 | Rv3670 Epoxide hydrolase (EC 3.3.2.9) |
| Rv2114 |
|
GO:0008853 | exodeoxyribonuclease III activity |
|
GO:0042586 | peptide deformylase activity |
|
GO:0040007 | growth |
|
GO:0001666 | response to hypoxia |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0008825 | cyclopropane-fatty-acyl-phospholipid synthase activity |
|
GO:0046500 | S-adenosylmethionine metabolic process |
|
GO:0052167 | modulation by symbiont of host innate immune response |
|
GO:0071766 | Actinobacterium-type cell wall biogenesis |
|
GO:0071768 | mycolic acid biosynthetic process |
|
GO:0071769 | mycolate cell wall layer assembly |
|
GO:0008444 | CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase activity |
|
GO:0006546 | glycine catabolic process |
|
GO:0005960 | glycine cleavage complex |
|
GO:0005618 | cell wall |
|
GO:0005887 | integral to plasma membrane |
|
GO:0004368 | glycerol-3-phosphate dehydrogenase activity |
|
GO:0004148 | dihydrolipoyl dehydrogenase activity |
|
GO:0003955 | NAD(P)H dehydrogenase (quinone) activity |
|
GO:0009405 | pathogenesis |
|
GO:0016655 | oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor |
|
GO:0050660 | flavin adenine dinucleotide binding |
|
GO:0051289 | protein homotetramerization |
|
GO:0055114 | oxidation-reduction process |
|
GO:0070401 | NADP+ binding |
|
GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD or NADP as acceptor |
|
GO:0005576 | extracellular region |



















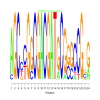
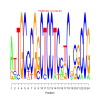
Discussion