Module 325 Residual: 0.54
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0325 | v02 | 0.54 | -12.67 |
| Rv0421c | Rv0423c Hydroxymethylpyrimidine phosphate synthase ThiC |
| Rv2392 Phosphoadenylyl-sulfate reductase [thioredoxin] (EC 1.8.4.8) / Adenylyl-sulfate reductase [thioredoxin] (EC 1.8.4.10) | Rv2715 Hydrolase, alpha/beta fold family |
| Rv2716 Phenazine biosynthesis protein PhzF like | Rv2773c Dihydrodipicolinate reductase (EC 1.3.1.26) |
| Rv3119 Molybdenum cofactor biosynthesis protein MoaE; Molybdopterin converting factor subunit 2 | Rv3120 Possible methyltransferase (EC 2.1.1.-) |
| Rv3487c putative esterase | Rv0422c Hydroxymethylpyrimidine phosphate kinase ThiD (EC 2.7.4.7) |
| Rv2391 Ferredoxin--sulfite reductase, actinobacterial type (EC 1.8.7.1) | Rv2393 Sirohydrochlorin ferrochelatase (EC 4.99.1.4) |
| Rv2968c PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN | Rv2969c POSSIBLE CONSERVED MEMBRANE OR SECRETED PROTEIN |
| Rv3825c Polyketide synthase | Rv3826 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0010467 | GO:0043170 | GO:0034645 | GO:0009059 | GO:0005840 | GO:0030529 | GO:0043228 | GO:0043232 | GO:0016779 | gene expression | macromolecule metabolic process | cellular macromolecule biosynthetic proc... | macromolecule biosynthetic process | ribosome | ribonucleoprotein complex | non-membrane-bounded organelle | intracellular non-membrane-bounded organ... | nucleotidyltransferase activity |
|
GO:0005829 | cytosol |
|
GO:0009228 | thiamine biosynthetic process |
|
GO:0005886 | plasma membrane |
|
GO:0040007 | growth |
|
GO:0043866 | adenylyl-sulfate reductase (thioredoxin) activity |
|
GO:0051539 | 4 iron, 4 sulfur cluster binding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0008839 | dihydrodipicolinate reductase activity |
|
GO:0019877 | diaminopimelate biosynthetic process |
|
GO:0051260 | protein homooligomerization |
|
GO:0070404 | NADH binding |
|
GO:0004091 | carboxylesterase activity |
|
GO:0004629 | phospholipase C activity |
|
GO:0010447 | response to acidity |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0052572 | response to host immune response |
|
GO:0008972 | phosphomethylpyrimidine kinase activity |
|
GO:0000103 | sulfate assimilation |
|
GO:0016002 | sulfite reductase activity |
|
GO:0051266 | sirohydrochlorin ferrochelatase activity |
|
GO:0008201 | heparin binding |
|
GO:0051701 | interaction with host |
|
GO:0009986 | cell surface |
|
GO:0005618 | cell wall |
|
GO:0006633 | fatty acid biosynthetic process |
|
GO:0010106 | cellular response to iron ion starvation |
|
GO:0004467 | long-chain fatty acid-CoA ligase activity |
|
GO:0046506 | sulfolipid biosynthetic process |


















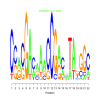
Discussion