Module 34 Residual: 0.54
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0034 | v02 | 0.54 | -13.86 |
| Rv1693 | Rv1694 RNA binding methyltransferase FtsJ like |
| Rv1695 NAD kinase (EC 2.7.1.23) | Rv2207 Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase (EC 2.4.2.21) |
| Rv2208 Cobalamin synthase | Rv2609c FIG019327: Probable conserved membrane protein |
| Rv2610c Phosphatidylinositol alpha-mannosyltransferase (EC 2.4.1.57) | Rv2611c Lauroyl/myristoyl acyltransferase involved in lipid A biosynthesis (Lauroyl/myristoyl acyltransferase) |
| Rv2612c CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase (EC 2.7.8.5) | Rv2613c FIG049476: HIT family protein |
| Rv2614c Threonyl-tRNA synthetase (EC 6.1.1.3) | Rv2854 Lysophospholipase (EC 3.1.1.5) |
| Rv2855 NADPH-dependent mycothiol reductase Mtr | Rv3806c POSSIBLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| Rv3807c FIG008913: Membrane-associated phospholipid phosphatase | Rv3808c Galactofuranosyl transferase (EC 2.-.-.-) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0016780 | phosphotransferase activity, for other s... |
|
GO:0003951 | NAD+ kinase activity |
|
GO:0006741 | NADP biosynthetic process |
|
GO:0006797 | polyphosphate metabolic process |
|
GO:0040007 | growth |
|
GO:0051289 | protein homotetramerization |
|
GO:0008939 | nicotinate-nucleotide-dimethylbenzimidazole phosphoribosyltransferase activity |
|
GO:0008818 | cobalamin 5'-phosphate synthase activity |
|
GO:0009236 | cobalamin biosynthetic process |
|
GO:0005618 | cell wall |
|
GO:0005886 | plasma membrane |
|
GO:0000030 | mannosyltransferase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0043750 | phosphatidylinositol alpha-mannosyltransferase activity |
|
GO:0008444 | CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase activity |
|
GO:0046474 | glycerophospholipid biosynthetic process |
|
GO:0004829 | threonine-tRNA ligase activity |
|
GO:0005829 | cytosol |
|
GO:0042803 | protein homodimerization activity |
|
GO:0050627 | mycothione reductase activity |
|
GO:0050660 | flavin adenine dinucleotide binding |
|
GO:0055114 | oxidation-reduction process |
|
GO:0070402 | NADPH binding |
|
GO:0005887 | integral to plasma membrane |
|
GO:0009247 | glycolipid biosynthetic process |
|
GO:0016763 | transferase activity, transferring pentosyl groups |
|
GO:0008921 | lipopolysaccharide-1,6-galactosyltransferase activity |
|
GO:0035250 | UDP-galactosyltransferase activity |
|
GO:0035496 | lipopolysaccharide-1,5-galactosyltransferase activity |
|
GO:0070592 | cell wall polysaccharide biosynthetic process |
|
GO:0071769 | mycolate cell wall layer assembly |


















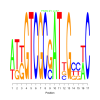

Discussion