Module 397 Residual: 0.58
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0397 | v02 | 0.58 | -19.55 |
| Rv1419 | Rv1977 Zn-dependent protease with chaperone function |
| Rv0270 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) | Rv0272c |
| Rv0273c Transcriptional regulator, TetR family | Rv0274 Glyoxalase family protein |
| Rv0389 Phosphoribosylglycinamide formyltransferase 2 (EC 2.1.2.-) | Rv0390 Rhodanese-related sulfurtransferase |
| Rv0676c Transmembrane transport protein MmpL5 | Rv0677c membrane protein, MmpS family |
| Rv0678 | Rv0834c PE-PGRS FAMILY PROTEIN |
| Rv0937c Ku domain protein | Rv0938 ATP-dependent DNA ligase (EC 6.5.1.1) clustered with Ku protein, LigD |
| Rv0939 Fumarylacetoacetase (EC 3.7.1.2) | Rv1989c |
| Rv1990c | Rv3917c Chromosome (plasmid) partitioning protein ParB / Stage 0 sporulation protein J |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006807 | GO:0016835 | nitrogen compound metabolic process | carbon-oxygen lyase activity |
|
GO:0005576 | extracellular region |
|
GO:0005886 | plasma membrane |
|
GO:0005829 | cytosol |
|
GO:0005887 | integral to plasma membrane |
|
GO:0006355 | regulation of transcription, DNA-dependent |
|
GO:0003690 | double-stranded DNA binding |
|
GO:0005515 | protein binding |
|
GO:0006303 | double-strand break repair via nonhomologous end joining |
|
GO:0033202 | DNA helicase complex |
|
GO:0042803 | protein homodimerization activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0003887 | DNA-directed DNA polymerase activity |
|
GO:0003896 | DNA primase activity |
|
GO:0003899 | DNA-directed RNA polymerase activity |
|
GO:0003909 | DNA ligase activity |
|
GO:0003910 | DNA ligase (ATP) activity |
|
GO:0004652 | polynucleotide adenylyltransferase activity |
|
GO:0005524 | ATP binding |
|
GO:0006269 | DNA replication, synthesis of RNA primer |
|
GO:0008310 | single-stranded DNA specific 3'-5' exodeoxyribonuclease activity |
|
GO:0030145 | manganese ion binding |
|
GO:0004334 | fumarylacetoacetase activity |
|
GO:0008150 | biological_process |
|
GO:0005694 | chromosome |
|
GO:0040007 | growth |




















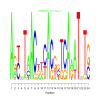
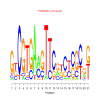
Discussion