Module 456 Residual: 0.56
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0456 | v02 | 0.56 | -7.54 |
| Rv0402c Transmembrane transport protein MmpL1 | Rv0404 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) |
| Rv0015c Serine/threonine-protein kinase PknA (EC 2.7.11.1) | Rv0016c Cell division protein FtsI [Peptidoglycan synthetase] (EC 2.4.1.129) |
| Rv0017c Cell division protein FtsW | Rv0403c membrane protein, MmpS family |
| Rv0405 Malonyl CoA-acyl carrier protein transacylase (EC 2.3.1.39) | Rv0424c |
| Rv0425c Metal cation transporting P-type ATPase CtpH | Rv1165 GTP-binding protein TypA/BipA |
| Rv1215c Cocaine esterase (EC 3.1.1.-) | Rv1607 Calcium/proton antiporter |
| Rv2751 O-Methyltransferase involved in polyketide biosynthesis | Rv3209 putative membrane protein |
| Rv3437 POSSIBLE CONSERVED TRANSMEMBRANE PROTEIN | Rv3438 |
|
GO:0005576 | extracellular region |
|
GO:0005886 | plasma membrane |
|
GO:0004467 | long-chain fatty acid-CoA ligase activity |
|
GO:0040007 | growth |
|
GO:0070566 | adenylyltransferase activity |
|
GO:0004672 | protein kinase activity |
|
GO:0004674 | protein serine/threonine kinase activity |
|
GO:0005515 | protein binding |
|
GO:0005829 | cytosol |
|
GO:0006468 | protein phosphorylation |
|
GO:0008360 | regulation of cell shape |
|
GO:0043085 | positive regulation of catalytic activity |
|
GO:0043086 | negative regulation of catalytic activity |
|
GO:0043388 | positive regulation of DNA binding |
|
GO:0046777 | protein autophosphorylation |
|
GO:0051055 | negative regulation of lipid biosynthetic process |
|
GO:0008955 | peptidoglycan glycosyltransferase activity |
|
GO:0008658 | penicillin binding |
|
GO:0030428 | cell septum |
|
GO:0008150 | biological_process |
|
GO:0008610 | lipid biosynthetic process |
|
GO:0009405 | pathogenesis |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0005525 | GTP binding |
|
GO:0006412 | translation |


















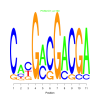

Discussion