Module 585 Residual: 0.58
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0585 | v02 | 0.58 | -10.81 |
| Rv2574 | Rv0092 Lead, cadmium, zinc and mercury transporting ATPase (EC 3.6.3.3) (EC 3.6.3.5); Copper-translocating P-type ATPase (EC 3.6.3.4) |
| Rv0314c Cell division protein DivIC (FtsB), stabilizes FtsL against RasP cleavage | Rv0337c Alanine transaminase (EC 2.6.1.2) |
| Rv1074c 3-ketoacyl-CoA thiolase (EC 2.3.1.16) @ Acetyl-CoA acetyltransferase (EC 2.3.1.9) | Rv1932 Thiol peroxidase, Tpx-type (EC 1.11.1.15) |
| Rv2046 Probable lipoprotein lppI | Rv2054 Dienelactone hydrolase family protein |
| Rv2259 Formaldehyde dehydrogenase MscR, NAD/mycothiol-dependent (EC 1.2.1.66) / S-nitrosomycothiol reductase MscR | Rv2260 Putative hydrolase in cluster with formaldehyde/S-nitrosomycothiol reductase MscR |
| Rv2482c Glycerol-3-phosphate acyltransferase (EC 2.3.1.15) | Rv2483c Phosphoserine phosphatase (EC 3.1.3.3) / 1-acyl-sn-glycerol-3-phosphate acyltransferase (EC 2.3.1.51) |
| Rv2484c Acyltransferase, ws/dgat/mgat subfamily protein | Rv3035 |
| Rv3042c Phosphoserine phosphatase (EC 3.1.3.3) | Rv3732 |
| Rv3853 Ribonuclease E inhibitor RraA |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0051188 | GO:0006732 | GO:0051186 | cofactor biosynthetic process | coenzyme metabolic process | cofactor metabolic process |
|
GO:0005886 | plasma membrane |
|
GO:0004008 | copper-exporting ATPase activity |
|
GO:0040007 | growth |
|
GO:0005576 | extracellular region |
|
GO:0004601 | peroxidase activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0051920 | peroxiredoxin activity |
|
GO:0055114 | oxidation-reduction process |
|
GO:0070402 | NADPH binding |
|
GO:0005829 | cytosol |
|
GO:0010127 | mycothiol-dependent detoxification |
|
GO:0016491 | oxidoreductase activity |
|
GO:0050607 | mycothiol-dependent formaldehyde dehydrogenase activity |
|
GO:0004416 | hydroxyacylglutathione hydrolase activity |
|
GO:0004366 | glycerol-3-phosphate O-acyltransferase activity |
|
GO:0004647 | phosphoserine phosphatase activity |
|
GO:0005887 | integral to plasma membrane |
|
GO:0070207 | protein homotrimerization |
|
GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity |



















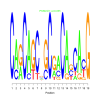
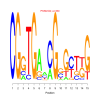
Discussion