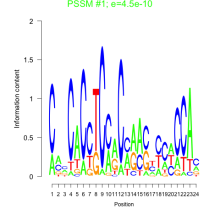
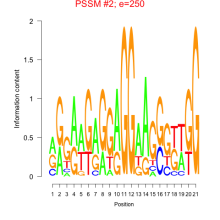
Thaps_bicluster_0151 Residual: 0.31
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0151 | 0.31 | Thalassiosira pseudonana |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
12982 AAA superfamily
14109 SrmB
15472 PP2Cc
19034 HSP70_NBD
20911 RNA14
21460 Peptidase_C19 superfamily
24445 hypothetical protein
25321 hypothetical protein
2569 Methyltransf_32 superfamily
25795 REX4_like
261747 PRK00142
262858 (IKI3) IKI3
263363 ANK
263786 (ZFP20) zf-TAZ
3009 hypothetical protein
31404 XRN1
33252 SrmB
34782 HrpA
35510 SET
38136 RRM2_PHIP1
38827 DEXDc
40402 SrmB
595 S1_IF1A
9061 RNA_pol_B_RPB2 superfamily
9576 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008152 | metabolism | 0.546944 | 1 | Thaps_bicluster_0151 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0473882 | 1 | Thaps_bicluster_0151 |
| GO:0006508 | proteolysis and peptidolysis | 0.424754 | 1 | Thaps_bicluster_0151 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.107561 | 1 | Thaps_bicluster_0151 |
| GO:0006396 | RNA processing | 0.102965 | 1 | Thaps_bicluster_0151 |
| GO:0006413 | translational initiation | 0.0438152 | 1 | Thaps_bicluster_0151 |
| GO:0006810 | transport | 0.294845 | 1 | Thaps_bicluster_0151 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.0910605 | 1 | Thaps_bicluster_0151 |

Comments