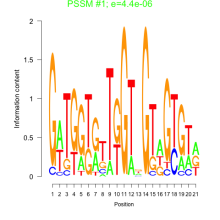
Thaps_bicluster_0279 Residual: 0.39
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0279 | 0.39 | Thalassiosira pseudonana |
Displaying 1 - 36 of 36
" class="views-fluidgrid-wrapper clear-block">
11220 eIF2A
1432 hypothetical protein
15950 Peptidase_A22B superfamily
20601 Lactamase_B superfamily
20621 hypothetical protein
21258 DUF3523
21710 Pho88 superfamily
22011 PqqL
22293 PLN00178
22770 PLN02908
23527 hypothetical protein
23655 hypothetical protein
26031 SHMT
261750 GckA
262136 Tim44 superfamily
262540 COG0523
262630 dnaj-like protein
264295 PP2Cc
264365 PRK08661
269240 (dnaK) dnaK
269662 Peptidase_M14_like superfamily
270365 hypothetical protein
28682 TCP1_theta
31810 pyruv_kin
32140 DHDPS
35049 PRK07411
40233 (Tp_CSF3) CSP_CDS
40771 (ENO2) PLN00191
4111 TENA_THI-4 superfamily
42612 pdxS
42704 (LEU2) IPMI
4487 hypothetical protein
6389 PDCD2_C superfamily
7558 DUF525 superfamily
7714 DUF1014
9705 DUF2431 superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006433 | prolyl-tRNA aminoacylation | 0.00454827 | 1 | Thaps_bicluster_0279 |
| GO:0051339 | regulation of lyase activity | 0.00454827 | 1 | Thaps_bicluster_0279 |
| GO:0006435 | threonyl-tRNA aminoacylation | 0.0090768 | 1 | Thaps_bicluster_0279 |
| GO:0006096 | glycolysis | 0.0119459 | 1 | Thaps_bicluster_0279 |
| GO:0006563 | L-serine metabolism | 0.0135857 | 1 | Thaps_bicluster_0279 |
| GO:0009098 | leucine biosynthesis | 0.0135857 | 1 | Thaps_bicluster_0279 |
| GO:0006915 | apoptosis | 0.0180749 | 1 | Thaps_bicluster_0279 |
| GO:0006544 | glycine metabolism | 0.0180749 | 1 | Thaps_bicluster_0279 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.0271022 | 1 | Thaps_bicluster_0279 |
| GO:0044267 | cellular protein metabolism | 0.061908 | 1 | Thaps_bicluster_0279 |


Comments