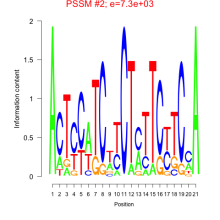
Thaps_bicluster_0048 Residual: 0.39
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0048 | 0.39 | Thalassiosira pseudonana |
Displaying 1 - 32 of 32
" class="views-fluidgrid-wrapper clear-block">
1015 hypothetical protein
11320 (DPHA1) PLN02291
11586 hypothetical protein
11756 SURF1 superfamily
12052 hypothetical protein
20959 (Tp_Myb1R4) regulator [Rayko]
22645 hypothetical protein
22940 ARGLU
23473 Cupin_8 superfamily
24311 hypothetical protein
2515 nst superfamily
25775 RING
260841 CysJ
261045 PI3Kc_like superfamily
261132 (PDT1) PLN02517 superfamily
261966 Ndh
264016 ATPase-IIB_Ca
264674 2OG-FeII_Oxy_3 superfamily
269113 M3_like
269735 hypothetical protein
30400 COG0390
38095 UPF0014 superfamily
3811 hypothetical protein
41521 Peptidase_S10
42320 PTZ00464 superfamily
5319 DPPIV_N
6597 hypothetical protein
6605 hypothetical protein
7840 hypothetical protein
8051 PLN02517 superfamily
9174 DUF563
9908 PRK14902
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006629 | lipid metabolism | 0.00347268 | 1 | Thaps_bicluster_0048 |
| GO:0015780 | nucleotide-sugar transport | 0.0154159 | 1 | Thaps_bicluster_0048 |
| GO:0006816 | calcium ion transport | 0.0184724 | 1 | Thaps_bicluster_0048 |
| GO:0009073 | aromatic amino acid family biosynthesis | 0.02152 | 1 | Thaps_bicluster_0048 |
| GO:0008643 | carbohydrate transport | 0.0426071 | 1 | Thaps_bicluster_0048 |
| GO:0006508 | proteolysis and peptidolysis | 0.0557851 | 1 | Thaps_bicluster_0048 |
| GO:0051341 | regulation of oxidoreductase activity | 0.0690961 | 1 | Thaps_bicluster_0048 |
| GO:0006306 | DNA methylation | 0.0863737 | 1 | Thaps_bicluster_0048 |
| GO:0006812 | cation transport | 0.125531 | 1 | Thaps_bicluster_0048 |
| GO:0016567 | protein ubiquitination | 0.219149 | 1 | Thaps_bicluster_0048 |


Comments