Regulatory Modules
Displaying 791 - 800 of 1058Desulfovibrio vulgaris Hildenborough
| Title | Motif 1 | Motif 2 | Residual | Bicluster Genes | Selected GO Terms |
|---|---|---|---|---|---|
| bicluster_0124 |
10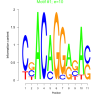 |
6200 |
0.41 | DVU0658, DVU1766, DVU2601, DVU2758, DVU2777, DVU3257, DVU0430, DVU0894, DVU1052, DVU1208, DVU2139, DVU2640, DVU3169, DVUA0148 | small molecule biosynthetic process, aromatic amino acid family metabolic pro..., aromatic amino acid family biosynthetic ..., aromatic compound biosynthetic process, organonitrogen compound biosynthetic pro... |
| bicluster_0125 |
21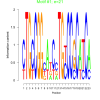 |
4.9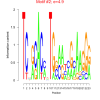 |
0.41 | DVU0310, DVU0311, DVU0312, DVU0313, DVU0314, DVU0315, DVU0512, DVU0513, DVU0515, DVU0516, DVU0517, DVU0519, DVU0913, DVU2232, DVU3003, DVU3018, DVU3019, DVU3020, DVU3004 | organelle organization, cell projection assembly, bacterial-type flagellum assembly, organelle assembly, cell projection organization |
| bicluster_0129 |
1000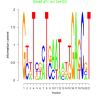 |
2100 |
0.40 | DVU1496, DVU1498, DVU1499, DVU1500, DVU1502, DVU1514, DVU1522, DVU1524, DVU1493, DVU1513, DVU1515, DVU1516, DVU1517, DVU1525 | DNA (cytosine-5-)-methyltransferase acti..., DNA-methyltransferase activity, S-adenosylmethionine-dependent methyltra..., methyltransferase activity, transferase activity, transferring one-c... |
| bicluster_0130 |
1900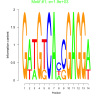 |
2500 |
0.38 | DVU0365, DVU1068, DVU1829, DVU2159, DVU2161, DVU2266, DVUA0009, DVU0375, DVU0497, DVU0887, DVU1490, DVU3129 | nitrogen cycle metabolic process, hydrolase activity, acting on glycosyl b... |
| bicluster_0132 |
3900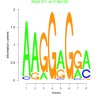 |
5800 |
0.48 | DVU0006, DVU0138, DVU0238, DVU0305, DVU0629, DVU0986, DVU0987, DVU1093, DVU1470, DVU1568, DVU1817, DVU1894, DVU1904, DVU2076, DVU2351, DVU2625, DVU2973, DVU0042, DVU0754, DVU0938, DVU0974, DVU1180, DVU2202, DVU2212 | electron carrier activity, protein methyltransferase activity, peptidyl-prolyl cis-trans isomerase acti..., cis-trans isomerase activity |
| bicluster_0133 |
0.00000092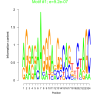 |
0.00037 |
0.48 | DVU0529, DVU0530, DVU0531, DVU0532, DVU0535, DVU0536, DVU0878, DVU0926, DVU0929, DVU1505, DVU1506, DVU2204, DVU2814, DVU2815, DVU2816, DVU2817, DVU2818, DVU2819, DVU0930, DVU1755 | lipid binding, endonuclease activity |
| bicluster_0135 |
1.9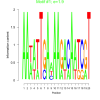 |
5000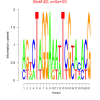 |
0.47 | DVU0299, DVU0300, DVU0570, DVU0677, DVU1440, DVU1532, DVU1548, DVU1606, DVU2251, DVU2747, DVU2933, DVU3016, DVU0079, DVU0328, DVU0691, DVU1004, DVU2373, DVU2609, DVU2779, DVU3127, DVU3392 | cell envelope organization, Gram-negative-bacterium-type cell outer ..., membrane biogenesis, single-organism membrane organization, membrane organization |
| bicluster_0143 |
0.0079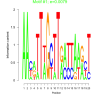 |
0.000047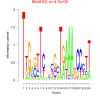 |
0.48 | DVU0111, DVU0459, DVU1144, DVU1175, DVU1509, DVU1758, DVU1834, DVU1849, DVU2415, DVU2912, DVU3084, DVU3089, DVU3328, DVU0880, DVU1285, DVU1664, DVU1665, DVU2300, DVU2340, DVU2655, DVU3054, DVU3150, DVU3240 | phosphorelay signal transduction system, signal transduction, signaling, intracellular signal transduction, single organism signaling |
| bicluster_0146 |
0.051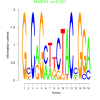 |
81000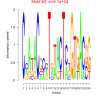 |
0.50 | DVU0841, DVU0899, DVU0936, DVU0965, DVU0966, DVU1038, DVU1067, DVU1238, DVU1392, DVU1615, DVU1910, DVU1990, DVU2150, DVU2364, DVU2783, DVU2940, DVU3171, DVU3187, DVU3238, DVU3277, DVU0041, DVU0896, DVU1064, DVU1406, DVU1423, DVU1954, DVU2056, DVU2251, DVU2782, DVU2791 | intramolecular oxidoreductase activity, intramolecular oxidoreductase activity, ..., NADP binding |
| bicluster_0148 |
0.029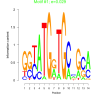 |
2.7 |
0.45 | DVU0174, DVU0220, DVU0253, DVU0278, DVU1569, DVU1816, DVU2202, DVU2557, DVU2968, DVU3263, DVU0006, DVU0138, DVU0238, DVU0305, DVU0883, DVU1568, DVU1812, DVU1904, DVU2076, DVU2583, DVU2616, DVU2684, DVU2935, DVU3227, DVU3296 | cellular homeostasis, homeostatic process, regulation of biological quality, biological regulation, copper ion binding |

