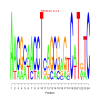
Module 255 Residual: 0.52
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0255 | v02 | 0.52 | -12.73 |
| Rv0247c Succinate dehydrogenase iron-sulfur protein (EC 1.3.99.1) | Rv0248c Succinate dehydrogenase flavoprotein subunit (EC 1.3.99.1) |
| Rv0632c Enoyl-CoA hydratase | Rv2951c Phthiodiolone/phenolphthiodiolone dimycocerosates ketoreductase |
| Rv2952 Phthiotriol/phenolphthiotriol dimycocerosates methyltransferase | Rv2971 2,5-diketo-D-gluconic acid reductase (EC 1.1.1.-) |
| Rv3464 dTDP-glucose 4,6-dehydratase (EC 4.2.1.46) | Rv3465 dTDP-4-dehydrorhamnose 3,5-epimerase (EC 5.1.3.13) |
| Rv3846 Superoxide dismutase [Mn/Fe] (EC 1.15.1.1) | Rv0164 |
| Rv1099c Fructose-1,6-bisphosphatase, GlpX type (EC 3.1.3.11) | Rv1436 NAD-dependent glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12) |
| Rv1437 Phosphoglycerate kinase (EC 2.7.2.3) | Rv1438 Triosephosphate isomerase (EC 5.3.1.1) |
| Rv2238c Alkyl hydroperoxide reductase subunit C-like protein | Rv2239c |
| Rv2970c lipase-esterase (lipN) |
|
GO:0000104 | succinate dehydrogenase activity |
|
GO:0005886 | plasma membrane |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0008171 | O-methyltransferase activity |
|
GO:0071770 | DIM/DIP cell wall layer assembly |
|
GO:0016491 | oxidoreductase activity |
|
GO:0040007 | growth |
|
GO:0008460 | dTDP-glucose 4,6-dehydratase activity |
|
GO:0008830 | dTDP-4-dehydrorhamnose 3,5-epimerase activity |
|
GO:0019305 | dTDP-rhamnose biosynthetic process |
|
GO:0042803 | protein homodimerization activity |
|
GO:0004784 | superoxide dismutase activity |
|
GO:0005506 | iron ion binding |
|
GO:0005576 | extracellular region |
|
GO:0006979 | response to oxidative stress |
|
GO:0009405 | pathogenesis |
|
GO:0051289 | protein homotetramerization |
|
GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity |
|
GO:0006094 | gluconeogenesis |
|
GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity |
|
GO:0004618 | phosphoglycerate kinase activity |
|
GO:0004807 | triose-phosphate isomerase activity |
|
GO:0004601 | peroxidase activity |
|
GO:0051260 | protein homooligomerization |



















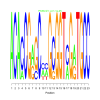
Discussion