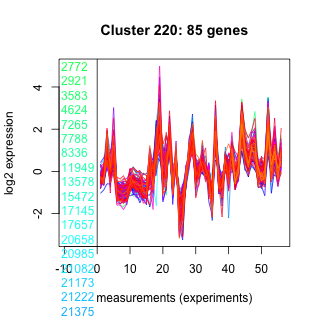
Thaps_hclust_0220 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009113 | purine base biosynthesis | 0.0103113 | 1 | Thaps_hclust_0220 |
| GO:0007046 | ribosome biogenesis | 0.0205206 | 1 | Thaps_hclust_0220 |
| GO:0006268 | DNA unwinding | 0.0255872 | 1 | Thaps_hclust_0220 |
| GO:0000059 | protein-nucleus import, docking | 0.0255872 | 1 | Thaps_hclust_0220 |
| GO:0009116 | nucleoside metabolism | 0.0306286 | 1 | Thaps_hclust_0220 |
| GO:0006396 | RNA processing | 0.0355342 | 1 | Thaps_hclust_0220 |
| GO:0042254 | ribosome biogenesis and assembly | 0.035645 | 1 | Thaps_hclust_0220 |
| GO:0016071 | mRNA metabolism | 0.0456032 | 1 | Thaps_hclust_0220 |
| GO:0006265 | DNA topological change | 0.0505452 | 1 | Thaps_hclust_0220 |
| GO:0006259 | DNA metabolism | 0.0700686 | 1 | Thaps_hclust_0220 |
|
2772 : Utp14 |
22404 : DEXDc superfamily |
30833 : Bystin |
40985 : Nucleostemin_like |
|
2921 : RRM_SF |
22448 : hypothetical protein |
31037 : DEADc |
42282 : HA2 |
|
3583 : PRK13796 |
22467 : AdoMet_MTases superfamily |
32374 : TPR |
42594 : Ribosomal_P0_like |
|
4624 : SrmB |
22582 : S1 |
32807 : DEADc |
42599 : SrmB |
|
7265 : PIN_SF superfamily |
22672 : hypothetical protein |
33088 : Obg_CgtA |
262275 : hypothetical protein |
|
7788 : (Tp_CCHH23) regulator [Rayko] |
22901 : hypothetical protein |
33425 : BMS1 |
262326 : ran binding protein 4-like protein |
|
8336 : hypothetical protein |
23187 : SDA1 |
34006 : Sun |
262722 : DEXDc superfamily |
|
11949 : Nop14 |
23202 : RsuA |
34027 : SrmB |
264345 : SpoU |
|
13578 : SrmB |
23245 : hypothetical protein |
36434 : DUF1253 |
268130 : hypothetical protein |
|
15472 : PP2Cc |
23650 : hypothetical protein |
36609 : PRK00142 |
268252 : Utp21 superfamily |
|
17145 : WBS_methylT |
23862 : Syo1_like superfamily |
36798 : S_TKc |
268331 : WD40 |
|
17657 : HSR1_MMR1 |
23900 : hypothetical protein |
37748 : SpoVK |
268559 : COG1084 |
|
20658 : Nop53 |
24016 : Ebp2 superfamily |
37965 : S1_like superfamily |
268598 : COG1094 |
|
20985 : COG5593 |
24369 : (Tp_CCCH1) RRM_SF superfamily |
38006 : FKBP_C |
268689 : RRM6_RBM19_RRM5_MRD1 |
|
21082 : Cupin_8 superfamily |
24590 : DUF663 |
38143 : WD40 superfamily |
268735 : (GPB1) WD40 |
|
21173 : hypothetical protein |
24712 : SrmB |
39075 : PRK09246 |
268766 : MdlB |
|
21222 : SrmB |
24849 : (Tp_CCCH8) DnaQ_like_exo superfamily |
39540 : Alpha_ANH_like_II |
269434 : COG1092 |
|
21375 : hypothetical protein |
25129 : hypothetical protein |
39845 : (DNA_TopoIV) PRK05560 |
269776 : Ribosomal_L1 |
|
22203 : (Tp_CCHH28) zf-C2H2_2 |
25183 : hypothetical protein |
39953 : SrmB |
bd1667 : (bd1667) Mpp10 |
|
22204 : hypothetical protein |
25484 : hypothetical protein |
40529 : COG2319 |
bd1515 : (bd1515) Adenylsuccinate_lyase_2 |
|
22212 : COG5638 |
29018 : NMD3 |
40871 : COG1444 |
bd1876 : (bd1876) NatB_MDM20 superfamily |
|
22334 : hypothetical protein |
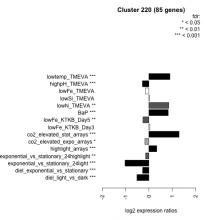
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | -0.512 | 0.000485 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.043 | 0.777 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | -0.255 | 0.00341 |
| BaP | BaP | 0.829 | 0.00037 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.142 | 0.00171 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 1.290 | 0.000658 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.902 | 0.000735 |
| highpH_TMEVA | highpH_TMEVA | -0.278 | 0.000725 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | -0.173 | 0.0218 |
| lowFe_TMEVA | lowFe_TMEVA | -0.154 | 0.452 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | -1.020 | 0.000581 |
| lowN_TMEVA | lowN_TMEVA | 0.851 | 0.00119 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | -0.271 | 0.000602 |
| lowSi_TMEVA | lowSi_TMEVA | 0.031 | 1 |
| highlight_arrays | highlight_arrays | 0.336 | 0.000442 |