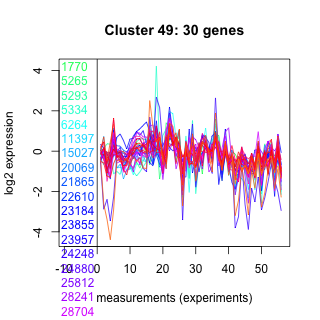
Thaps_hclust_0049 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006414 | translational elongation | 0.000000753 | 0.00274955 | Thaps_hclust_0049 |
| GO:0009052 | pentose-phosphate shunt, non-oxidative branch | 0.0131889 | 1 | Thaps_hclust_0049 |
| GO:0016089 | aromatic amino acid family biosynthesis, shikimate pathway | 0.0131889 | 1 | Thaps_hclust_0049 |
| GO:0006030 | chitin metabolic process | 0.0131889 | 1 | Thaps_hclust_0049 |
| GO:0019856 | pyrimidine base biosynthesis | 0.0197201 | 1 | Thaps_hclust_0049 |
| GO:0009098 | leucine biosynthesis | 0.0197201 | 1 | Thaps_hclust_0049 |
| GO:0006432 | phenylalanyl-tRNA aminoacylation | 0.0262093 | 1 | Thaps_hclust_0049 |
| GO:0005975 | carbohydrate metabolic process | 0.0326569 | 1 | Thaps_hclust_0049 |
| GO:0006281 | DNA repair | 0.0366287 | 1 | Thaps_hclust_0049 |
| GO:0042254 | ribosome biogenesis and assembly | 0.0454283 | 1 | Thaps_hclust_0049 |
|
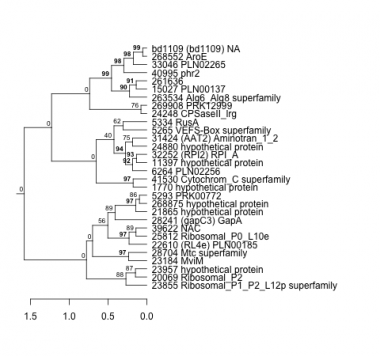
1770 : hypothetical protein |
21865 : hypothetical protein |
28241 : (gapC3) GapA |
41530 : Cytochrom_C superfamily |
|
5265 : VEFS-Box superfamily |
22610 : (RL4e) PLN00185 |
28704 : Mtc superfamily |
261636 : |
|
5293 : PRK00772 |
23184 : MviM |
31424 : (AAT2) Aminotran_1_2 |
263534 : Alg6_Alg8 superfamily |
|
5334 : RusA |
23855 : Ribosomal_P1_P2_L12p superfamily |
32252 : (RPI2) RPI_A |
268552 : AroE |
|
6264 : PLN02256 |
23957 : hypothetical protein |
33046 : PLN02265 |
268875 : hypothetical protein |
|
11397 : hypothetical protein |
24248 : CPSaseII_lrg |
39622 : NAC |
269908 : PRK12999 |
|
15027 : PLN00137 |
24880 : hypothetical protein |
40995 : phr2 |
bd1109 : (bd1109) NA |
|
20069 : Ribosomal_P2 |
25812 : Ribosomal_P0_L10e |
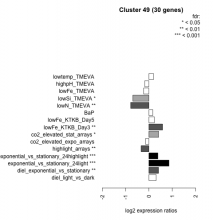
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | 0.311 | 0.0506 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.586 | 0.00161 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.238 | 0.141 |
| BaP | BaP | 0.166 | 0.368 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.386 | 0.000526 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.420 | 0.018 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.198 | 0.361 |
| highpH_TMEVA | highpH_TMEVA | -0.180 | 0.178 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | -0.144 | 0.38 |
| lowFe_TMEVA | lowFe_TMEVA | -0.225 | 0.37 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.860 | 0.000581 |
| lowN_TMEVA | lowN_TMEVA | -0.780 | 0.00119 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.413 | 0.00388 |
| lowSi_TMEVA | lowSi_TMEVA | -0.690 | 0.0296 |
| highlight_arrays | highlight_arrays | -0.335 | 0.00709 |