Module 482 Residual: 0.61
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0482 | v02 | 0.61 | -15.41 |
| Rv0490 Phosphate regulon sensor protein PhoR (SphS) (EC 2.7.13.3); Sensor-like histidine kinase senX3 (EC 2.7.13.3) | Rv0410c serine/threonine protein kinase |
| Rv0411c \Glutamine ABC transporter, periplasmic glutamine-binding protein (TC 3.A.1.3.2)\\\"" | Rv0412c Possible membrane protein |
| Rv0413 \Possible mutator protein MutT3 (7,8-dihydro-8-oxoguanine-triphosphatase) \\\"" | Rv0486 Glycosyltransferase MshA involved in mycothiol biosynthesis (EC 2.4.1.-) |
| Rv0487 Uncharacterized protein Rv0487/MT0505 clustered with mycothiol biosynthesis gene | Rv1330c Nicotinate phosphoribosyltransferase (EC 2.4.2.11) |
| Rv1331 ATP-dependent Clp protease adaptor protein ClpS | Rv1332 Transcriptional regulatory protein |
| Rv2130c L-cysteine:1D-myo-inosityl 2-amino-2-deoxy-alpha-D-glucopyranoside ligase MshC | Rv2131c Protein cysQ homolog |
| Rv2234 Low molecular weight protein tyrosine phosphatase (EC 3.1.3.48) | Rv2235 Cytochrome oxidase biogenesis protein Surf1, facilitates heme A insertion |
| Rv2358 Transcriptional regulator, ArsR family | Rv2359 Zinc uptake regulation protein ZUR |
| Rv2372c Ribosomal RNA small subunit methyltransferase E (EC 2.1.1.-) | Rv2583c GTP pyrophosphokinase (EC 2.7.6.5), (p)ppGpp synthetase I |
| Rv2589 Gamma-aminobutyrate:alpha-ketoglutarate aminotransferase (EC 2.6.1.19) |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0034645 | GO:0009059 | GO:0043170 | GO:0005737 | cellular macromolecule biosynthetic proc... | macromolecule biosynthetic process | macromolecule metabolic process | cytoplasm |
|
GO:0008150 | biological_process |
|
GO:0005576 | extracellular region |
|
GO:0009405 | pathogenesis |
|
GO:0018106 | peptidyl-histidine phosphorylation |
|
GO:0046777 | protein autophosphorylation |
|
GO:0001899 | negative regulation of cytolysis by symbiont of host cells |
|
GO:0004674 | protein serine/threonine kinase activity |
|
GO:0005515 | protein binding |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0006538 | glutamate catabolic process |
|
GO:0006543 | glutamine catabolic process |
|
GO:0040007 | growth |
|
GO:0044413 | avoidance of host defenses |
|
GO:0052027 | modulation by symbiont of host signal transduction pathway |
|
GO:0052190 | modulation by symbiont of host phagocytosis |
|
GO:0052369 | positive regulation by symbiont of defense-related host reactive oxygen species production |
|
GO:0005887 | integral to plasma membrane |
|
GO:0008375 | acetylglucosaminyltransferase activity |
|
GO:0010125 | mycothiol biosynthetic process |
|
GO:0004516 | nicotinate phosphoribosyltransferase activity |
|
GO:0009435 | NAD biosynthetic process |
|
GO:0034355 | NAD salvage |
|
GO:0047280 | nicotinamide phosphoribosyltransferase activity |
|
GO:0004817 | cysteine-tRNA ligase activity |
|
GO:0016049 | cell growth |
|
GO:0035446 | cysteine-glucosaminylinositol ligase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity |
|
GO:0008934 | inositol monophosphate 1-phosphatase activity |
|
GO:0030145 | manganese ion binding |
|
GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity |
|
GO:0042803 | protein homodimerization activity |
|
GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process |
|
GO:0050897 | cobalt ion binding |
|
GO:0004725 | protein tyrosine phosphatase activity |
|
GO:0006470 | protein dephosphorylation |
|
GO:0052046 | modification by symbiont of host morphology or physiology via secreted substance |
|
GO:0052083 | negative regulation by symbiont of host cell-mediated immune response |
|
GO:0004438 | phosphatidylinositol-3-phosphatase activity |
|
GO:0005618 | cell wall |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0010043 | response to zinc ion |
|
GO:0003700 | sequence-specific DNA binding transcription factor activity |
|
GO:0008270 | zinc ion binding |
|
GO:0045893 | positive regulation of transcription, DNA-dependent |
|
GO:0008728 | GTP diphosphokinase activity |
|
GO:0008893 | guanosine-3',5'-bis(diphosphate) 3'-diphosphatase activity |
|
GO:0015968 | stringent response |





















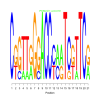

Discussion