Module 502 Residual: 0.52
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0502 | v02 | 0.52 | -25.82 |
| Rv2378c L-lysine 6-monooxygenase MbtG (EC 1.14.13.59) | Rv2379c Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) |
| Rv2380c Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) | Rv2381c Polyketide synthetase MbtD |
| Rv2382c Malonyl CoA-acyl carrier protein transacylase (EC 2.3.1.39) | Rv2383c Phenyloxazoline synthase MbtB (EC 6.3.2.-) |
| Rv2384 Bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase MbtA | Rv2386c isochorismate synthase MbtI |
| Rv0450c Transmembrane transport protein MmpL4 | Rv1342c CONSERVED MEMBRANE PROTEIN |
| Rv1343c Lipoprotein LprD | Rv1344 Acyl carrier protein MbtL |
| Rv1345 Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) | Rv1346 Isovaleryl-CoA dehydrogenase (EC 1.3.99.10) |
| Rv1347c Lysine N-acyltransferase mbtK (EC 2.3.1.-) (N-acyltransferase mbtK) (Mycobactin synthetase protein K) | Rv1348 ABC transporter ATP-binding protein |
| Rv1349 ABC transporter ATP-binding protein | Rv1845c Peptidase M48, Ste24p precursor |
| Rv1846c Transcriptional regulator, BlaI family | Rv1847 Putative esterase |
|
GO:0040007 | growth |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0000036 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process |
|
GO:0031177 | phosphopantetheine binding |
|
GO:0004314 | [acyl-carrier-protein] S-malonyltransferase activity |
|
GO:0009405 | pathogenesis |
|
GO:0010106 | cellular response to iron ion starvation |
|
GO:0019290 | siderophore biosynthetic process |
|
GO:0052572 | response to host immune response |
|
GO:0008668 | (2,3-dihydroxybenzoyl)adenylate synthase activity |
|
GO:0016881 | acid-amino acid ligase activity |
|
GO:0004049 | anthranilate synthase activity |
|
GO:0000287 | magnesium ion binding |
|
GO:0004106 | chorismate mutase activity |
|
GO:0008909 | isochorismate synthase activity |
|
GO:0009697 | salicylic acid biosynthetic process |
|
GO:0043904 | isochorismate pyruvate lyase activity |
|
GO:0005576 | extracellular region |
|
GO:0044119 | growth of symbiont in host cell |
|
GO:0005887 | integral to plasma membrane |
|
GO:0005515 | protein binding |
|
GO:0005524 | ATP binding |
|
GO:0008922 | long-chain fatty acid [acyl-carrier-protein] ligase activity |
|
GO:0004466 | long-chain-acyl-CoA dehydrogenase activity |
|
GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors |
|
GO:0008415 | NA |
|
GO:0016291 | NA |
|
GO:0016410 | N-acyltransferase activity |
|
GO:0034069 | aminoglycoside N-acetyltransferase activity |
|
GO:0015891 | siderophore transport |
|
GO:0042927 | siderophore transporter activity |
|
GO:0052099 | acquisition by symbiont of nutrients from host via siderophores |
|
GO:0055072 | iron ion homeostasis |
|
GO:0075139 | response to host iron concentration |
|
GO:0003677 | DNA binding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0006355 | regulation of transcription, DNA-dependent |
|
GO:0046677 | response to antibiotic |






















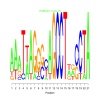
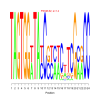
Discussion