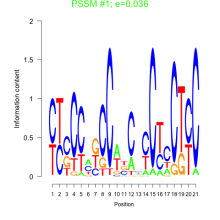
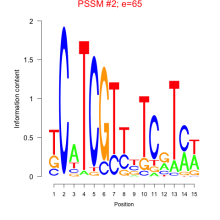
Thaps_bicluster_0013 Residual: 0.47
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0013 | 0.47 | Thalassiosira pseudonana |
Displaying 1 - 56 of 56
" class="views-fluidgrid-wrapper clear-block">
11454 C2B_Copine
11905 hypothetical protein
1428 hypothetical protein
14370 WD40 superfamily
1492 hypothetical protein
17140 AdoMet_MTases superfamily
17228 Tryp_SPc
18846 IFT46_B_C superfamily
20577 hypothetical protein
21200 hypothetical protein
21292 ArsB_NhaD_permease superfamily
21718 hypothetical protein
21727 DENN superfamily
21850 hypothetical protein
22337 hypothetical protein
23240 hypothetical protein
23247 choice_anch_B
23261 (Tp_fungal_TRF1a) regulator [Rayko]
23688 hypothetical protein
2399 Str_synth superfamily
24153 hypothetical protein
24187 hypothetical protein
24189 (Tp_bZIP14) regulator [Rayko]
24387 vWFA superfamily
24649 hypothetical protein
25050 ArfGap
25295 Gst
25393 hypothetical protein
261479 (Tp_E2F-DP1a_SET1a) DP superfamily
262099 PLPDE_III_AR
26224 SPFH_prohibitin
26398 Peptidase_C12_UCH37_BAP1
264431 Steroid_dh superfamily
268404 (Lig1) dnl1
270031 MhpC
270113 hypothetical protein
29183 PRK12344
2996 hypothetical protein
30921 MtN3_slv superfamily
32596 Tryp_SPc
3328 hypothetical protein
35690 CysH
35808 SOUL
4536 hypothetical protein
4609 hypothetical protein
4878 hypothetical protein
5916 rad3
6640 hypothetical protein
7630 hypothetical protein
8370 ANK
843 Rab1_Ypt1
8964 LRR_RI superfamily
909 hypothetical protein
9258 HP_PGM_like
9301 hypothetical protein
9551 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009401 | phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.0060732 | 1 | Thaps_bicluster_0013 |
| GO:0006522 | alanine metabolism | 0.00661567 | 1 | Thaps_bicluster_0013 |
| GO:0019379 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin) | 0.00661567 | 1 | Thaps_bicluster_0013 |
| GO:0019752 | carboxylic acid metabolism | 0.0131889 | 1 | Thaps_bicluster_0013 |
| GO:0006030 | chitin metabolic process | 0.0131889 | 1 | Thaps_bicluster_0013 |
| GO:0006614 | SRP-dependent cotranslational protein-membrane targeting | 0.0390631 | 1 | Thaps_bicluster_0013 |
| GO:0000074 | regulation of cell cycle | 0.0591753 | 1 | Thaps_bicluster_0013 |
| GO:0043087 | regulation of GTPase activity | 0.0642796 | 1 | Thaps_bicluster_0013 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 0.0948998 | 1 | Thaps_bicluster_0013 |
| GO:0006310 | DNA recombination | 0.118695 | 1 | Thaps_bicluster_0013 |

Comments