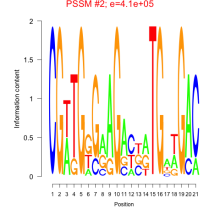
Thaps_bicluster_0095 Residual: 0.37
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0095 | 0.37 | Thalassiosira pseudonana |
Displaying 1 - 29 of 29
" class="views-fluidgrid-wrapper clear-block">
10803 (Tp_HSF_2.4c) regulator [Rayko]
10916 hypothetical protein
12403 SNF2_N
1457 hypothetical protein
17031 PI3Kc_like superfamily
20732 PKc_like superfamily
21538 PLN02666
21576 hypothetical protein
21676 DUF2252 superfamily
21752 hypothetical protein
22277 RdRP
22355 hypothetical protein
22932 (Tp_HSF_4.2f) HSF_DNA-bind
23562 hypothetical protein
23894 hypothetical protein
23988 DUF1800 superfamily
24231 C2 superfamily
25015 CAP10 superfamily
25332 (Tp_HSF_2.4g) HSF_DNA-bind superfamily
261472 WD40 superfamily
261678 CYCc
263403 YafJ
264222 POLBc superfamily
268351 (Tp_Hox2) regulator [Rayko]
269496 (Tp_TAZ3_PHD1) KAT11 superfamily
33039 SpoT
36301 HMG-box
40445 Dynein_heavy superfamily
9571 (Tp_HSF_3.2i) HSF_DNA-bind
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006355 | regulation of transcription, DNA-dependent | 0.00000601 | 0.0166013 | Thaps_bicluster_0095 |
| GO:0015969 | guanosine tetraphosphate metabolism | 0.0148069 | 1 | Thaps_bicluster_0095 |
| GO:0045449 | regulation of transcription | 0.0221325 | 1 | Thaps_bicluster_0095 |
| GO:0009190 | cyclic nucleotide biosynthesis | 0.0789128 | 1 | Thaps_bicluster_0095 |
| GO:0006260 | DNA replication | 0.122739 | 1 | Thaps_bicluster_0095 |
| GO:0007242 | intracellular signaling cascade | 0.145514 | 1 | Thaps_bicluster_0095 |
| GO:0007018 | microtubule-based movement | 0.148722 | 1 | Thaps_bicluster_0095 |
| GO:0016567 | protein ubiquitination | 0.25691 | 1 | Thaps_bicluster_0095 |
| GO:0006468 | protein amino acid phosphorylation | 0.556068 | 1 | Thaps_bicluster_0095 |
| GO:0006508 | proteolysis and peptidolysis | 0.669444 | 1 | Thaps_bicluster_0095 |


Comments